
Therapeutic Area Data Standards User Guide for Colorectal Cancer
Version 1.0 (Provisional)
Notes to Readers
- This is the provisional version 1.0 of the Therapeutic Area User Guide for Colorectal Cancer.
- This document is based on CDASH v1.1, CDASHUG v1.0, SDTM v1.4, SDTMIG v3.2, SDTMIG-PGx v1.0, SDTMIG-MD v1.0, ADaM v2.1, and ADaMIG v1.1.
Revision History
| Date | Version |
|---|---|
| 2018-11-15 | 1.0 Provisional |
© 2018 Clinical Data Interchange Standards Consortium, Inc. All rights reserved.
Contents
- 1 Introduction
- 2 Overview of Colorectal Cancer
- 3 Subject and Disease Characteristics
- 3.1 Initial Diagnosis
- 3.2 Staging
- 3.3 Pathology
- 4 Disease Assessments
- 4.1 Treatment
- 4.1.1 Anti-cancer Therapy
- 4.2 Disease Assessments and Response
- 4.1 Treatment
- 5 Questionnaires, Ratings, and Scales
- 6 Routine Data
- 7 Analysis Data
- Appendices
- Appendix A: Colorectal Cancer Team
- Appendix B: Non-Standard Variables (NSVs)
- Appendix C: Glossary and Abbreviations
- Appendix D: References
- Appendix E: Representations and Warranties, Limitations of Liability, and Disclaimers
1 Introduction
The Therapeutic Area Data Standards User Guide for Colorectal Cancer (TAUG-CrCa) was developed under the Coalition for Accelerating Standards and Therapies (CFAST) initiative.
The purpose of the TAUG-CrCa is to describe how to use CDISC standards to represent data pertaining to studies in colorectal cancer. TAUG-CrCa Version 1.0 focuses on clinical trials in metastatic colorectal cancer.
This document provides advice and examples for Clinical Data Acquisition Standards Harmonization (CDASH), the Study Data Tabulation Model (SDTM), the SDTM Implementation Guide for Medical Devices (SDTMIG-MD), and the Analysis Data Model (ADaM), including
-
guidance on the use of domains and variables;
-
annotated sample case report forms (CRFs) compliant with CDASH;
- examples of SDTM datasets, with text describing the situational context and pointing out records of note; and
- discussion of the concepts and common analysis endpoints relevant to the analysis of colorectal cancer studies.
The biomedical concepts covered in this guide were selected from concepts identified by one or more stakeholders as important, and which were not addressed or were not completely addressed by existing CDISC implementation guides. This TAUG does not provide guidance on what data are needed for regulatory submission or approval; it only provides advice on how to represent data in a standard form.
A biomedical concept is a unit of knowledge created by a unique combination of the characteristics that define observations of real-world, clinical research phenomena. A biomedical concept represents healthcare and/or clinical research knowledge that borrows from medical knowledge, statistical knowledge, and the Biomedical Research Integrated Domain Group (BRIDG) model. Metadata for biomedical concepts include the properties of the data items that are parts of the biomedical concepts, controlled terminology for those data items, and the ways in which the biomedical concepts relate to each other.
This user guide emphasizes that examples are only examples and should not be over-interpreted. For guidance on the selection of biomedical concepts and endpoints, please refer to the appropriate clinical and regulatory authorities. Clinical guidelines, articles, and other works consulted by the team during the creation of this document are referenced where appropriate, using the American Medical Association (AMA) style for citation. For a full list of references, see Appendix D: References.
1.1 How to Read this Document
- First, read foundational standards upon which this document is based: CDASH v1.1, CDASHUG v1.0, SDTM v1.4, SDTMIG v3.2, SDTMIG-PGx v1.0, SDTMIG-MD v1.0, ADaM v2.1, and ADaMIG v1.1 to gain some familiarity with data models and the basic rules for how they are implemented. These standards are available from: http://www.cdisc.org/.
- For guidance on how to review the CDASH annotated case report forms (aCRFs) that were created based on the CDASH metadata included with a CRF, see Section 1.3, CDASH Metadata and Annotated CRFs.
- Next, read Introduction to Therapeutic Area Standards (https://wiki.cdisc.org/x/SSy8AQ) to be sure to know what to expect from such a document.
- Read this guide all the way through (without skipping any sections) at least once.
- Consult any sections of particular interest as the need arises.
Some things to bear in mind while reading this TAUG:
- This TAUG does not replace or supersede the foundational CDISC standards or their implementation guides, and should not be used as a substitute for any other CDISC standard.
- This document does not repeat content already published in another CDISC standard.
- This document is not and does not try to be an exhaustive documentation of every possible kind of data that could be collected in relation to colorectal cancer. Instead, the team has tried to focus on those areas that CFAST's resources have identified as most likely to be relevant and useful more often than not.
- The advice and examples presented in this document are influenced by ongoing internal standards development at CDISC. If a modeling approach seems inconsistent with a published standard, it may be a genuine error, but it could also be a reflection of potential or upcoming changes to the standard.
- The examples in this document use CDISC Controlled Terminology where possible, but some values that seem to be controlled terminology may still be under development at the time of publication, or even especially plausible "best-guess" placeholder values. Do not rely on any source other than the CDISC value set in the NCI Thesaurus (available at http://www.cancer.gov/research/resources/terminology/cdisc) for controlled terminology.
- With time, some parts of this document may become outdated. Those parts will be updated in the next version.
1.2 Organization of this Document
This document is divided into the following sections:
- Section 1, Introduction, provides an overall introduction to the purpose and goals of the colorectal cancer project.
- Section 2, Overview of Colorectal Cancer, provides a brief overview of the focus of this document in relation to the stage of colorectal cancer.
- Section 3, Subject and Disease Characteristics, covers data that are usually collected once at the beginning of a study.
- Section 4, Disease Assessments, covers data that are used to evaluate disease severity, control, or progression. These are usually collected repeatedly during a study and may be used as, or for the derivation of, efficacy and/or safety endpoints.
- Section 5, Questionnaires, Ratings, and Scales
- Section 6, Routine Data, provides information on routine data collected particular to colorectal cancer.
- Section 7, Analysis Data, includes key data analysis concepts for a colorectal cancer study.
- Appendices provide additional background material and describe other supplemental material relevant to colorectal cancer.
1.3 CDASH Metadata and Annotated CRFs
CDASH examples include both metadata tables and sample case report forms (CRFs). Each table of CDASH metadata corresponds to a sample annotated CRF (aCRF), built directly from the metadata. The annotations show the variables associated with each field in the context of data collection (CDASH) and submission (SDTM). Which context is applicable is denoted by color. Data that are collected using the same variable name as defined in the SDTMIG are in RED. If the CDASHIG variable differs from the one defined in the SDTMIG, the CDASHIG variable is in GREY. Data collected, but not submitted in SDTM-based datasets, are denoted as NOT SUBMITTED.
The following diagram illustrates how to interpret the annotations.
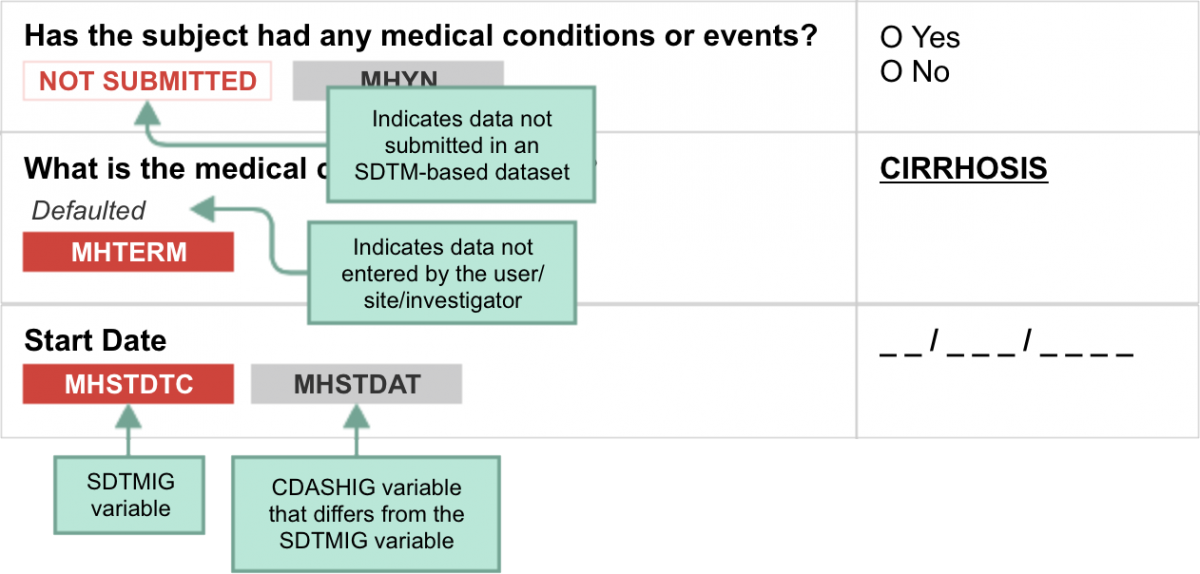
CDASH variables may also be mapped to or used to populate other SDTMIG variables that are not shown. The information in the attached CRF metadata has been "customized" following the conformance rules included in the CDASHIG to illustrate data collection instruments relevant to this TAUG. Users should always refer to the CDASHIG and CDASH Model when creating or adapting these CRFs to other studies.
When viewing sample aCRFs, bear in mind that:
- More information may also be found in the CDASH Model and CDASHIG.
- Example CRFs are provided to illustrate data collection instruments. They are only examples and are not meant to imply that any particular layout is preferable over another.
- Example CRFs are best understood in conjunction with their respective metadata tables and/or the CDASH Domain Metadata Tables.
- Most example CRFs do not include the Highly Recommended header variables. The population of these values is usually determined by the sponsor's data management system.
- Sponsors are responsible for understanding and implementing CDISC Controlled Terminology where applicable.
- CDASH variable names for denormalized variables are examples. Sponsor may use other conventions for creating denormalized CDASH variable names.
- CDASH variable names that are annotated as "NOT SUBMITTED" may be used to contribute towards the population of other appropriate variables when the SDTM-based datasets are created.
- CDASH variables may also be mapped to or used to populate other SDTMIG variables that are not shown.
1.4 Known Issues
Non-Standard Variables: Non-standard variables (NSVs) are shown as though they were appended to a dataset rather than being represented as supplemental qualifiers because this makes examples easier to understand. It is also consistent with a proposed future structure for representing NSVs. That structure is a modification of the NSV proposal that went for public review, and is still under development. For a list of all NSVs used in this document, and the variable-level metadata that might become normative for the NSVs should they be promoted to standard variables, see Appendix B: Non-Standard Variables (NSVs).
-
NSV Naming Convention: In this document, NSV names include the 2-letter domain code before the variable name. The naming convention for these variables is under discussion.
- Treatment Regimens: The strategy used to treat cancer is often a "regimen" that may consist of multiple drugs or of drug treatment combined with radiation and/or surgery. The SDTMIG does not give clear guidance on how to indicate that multiple treatment modalities are combined to create a regimen or a product. The modeling of this data is under consideration in the Combination Therapy Focus Area User Guide and will not be part of this TAUG.
- If Tumor Is Inevaluable, Reason Not Done (Target and Non-Target Lesions): Current modeling of tumor state of inevaluable shows that the result will be missing, status will be NOT DONE, and the reason will be mapped to the --REASND variable. This modeling does not capture that the tumor was inevaluable. At the time of publication of this document, there were ongoing discussions on how best to model this data; users are cautioned that the current modeling may be subject to change. The pre-specified terms used for --REASND are simply examples of collected terms to encourage standardized terminology (as opposed to free-text). Sponsors are encouraged to use generic reasons, and if necessary to provide any other detailed reasons in the Comments (CO) domain. The CDASH metadata tables provide suggestions on the use of these generic reasons.
- CDASH Variable Name, Question Text, and Prompt: The aCRFs were developed considering the CDASHIG v2.0 and CDASH Model v1.0 that are anticipated to be published. Thus, users are cautioned that the CRF prompts, question texts, and CDASH variable names used in this TAUG are subject to change.
- Collection of Date of Birth: In several countries, the collection of date of birth is restricted in order to protect patient confidentiality. There are some concepts (e.g., "Age at Diagnosis") that rely on the date of birth in order for that concept to be derived. Collection of age for these critical age-related data points has been raised to the CDISC Submission Data Standards Committee for discussion on the best way to represent this in the SDTM. Guidance on how to collect this information will be provided in future versions of this TAUG.
- Baseline Disease Characteristics and Identification of Primary Tumor: These concepts are being developed in the Lung Cancer TAUG (currently in development at the time of publication of this TAUG). Once modeling of these concepts has been approved they may be added to this TAUG in a future version.
- SDTMIG-PGx Recommendations: Genetic variations data were not parsed using PFORRESF, PFORRES and PFGENLOC. Although this parsing approach is recommended by the SDTMIG-PGx, the non-parsing approach was used to facilitate historical data collected on CRFs. This approach is under discussion within the PGx Team.
2 Overview of Colorectal Cancer
Colorectal cancer is a growth of malignant cells in the colon and/or rectum. Globally, colorectal cancer is the third most common cancer in men and the second in women.[1] Although important efforts in the prevention and early detection of colorectal cancer are ongoing, some patients present with metastases at initial diagnosis and many patients with non-metastatic colorectal cancer will develop metastases.
The concept map below provides a high-level overview of the stages of colorectal cancer, including the American Joint Committee on Cancer (AJCC) TNM staging system (the most commonly used colorectal cancer staging system). This system looks at T (tumor expansion), N (extent of cancer spread to lymph nodes) and M (metastases or spread of cancer to other organs). Also included is the transition from non-metastatic to metastatic colorectal cancer, where the cancer has spread through the colon wall and may have spread to nearby organs and/or lymph nodes. The content of this user guide has been developed in relation to Stage IV colorectal cancer. In Stage IVA, cancer has spread to 1 organ (e.g., liver, lung, ovary) or to a non-regional lymph node. In Stage IVB, cancer has spread to more than 1 organ/site or to the peritoneum (note that the peritoneum is not included in the Japanese classification of colorectal carcinoma) . Once cancer has spread to another part of the body it is unlikely to be curable. However, for some people with colorectal cancer that has only spread to the liver or lungs, a cure may still be possible.[2]
For readers unfamiliar with colorectal cancer, basic information in layman's language can be found at the following websites:
- U.S. National Library of Medicine (https://www.nlm.nih.gov/medlineplus/colorectalcancer.html)
- National Cancer Institute (https://www.cancer.gov)
-
Japanese Society for Cancer of the Colon and Rectum (JSCCR) Guidelines 2014 (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4653248/)
Concept Map. Overview of Non-metastatic and Metastatic Settings in Colorectal Cancer: Classification and Settings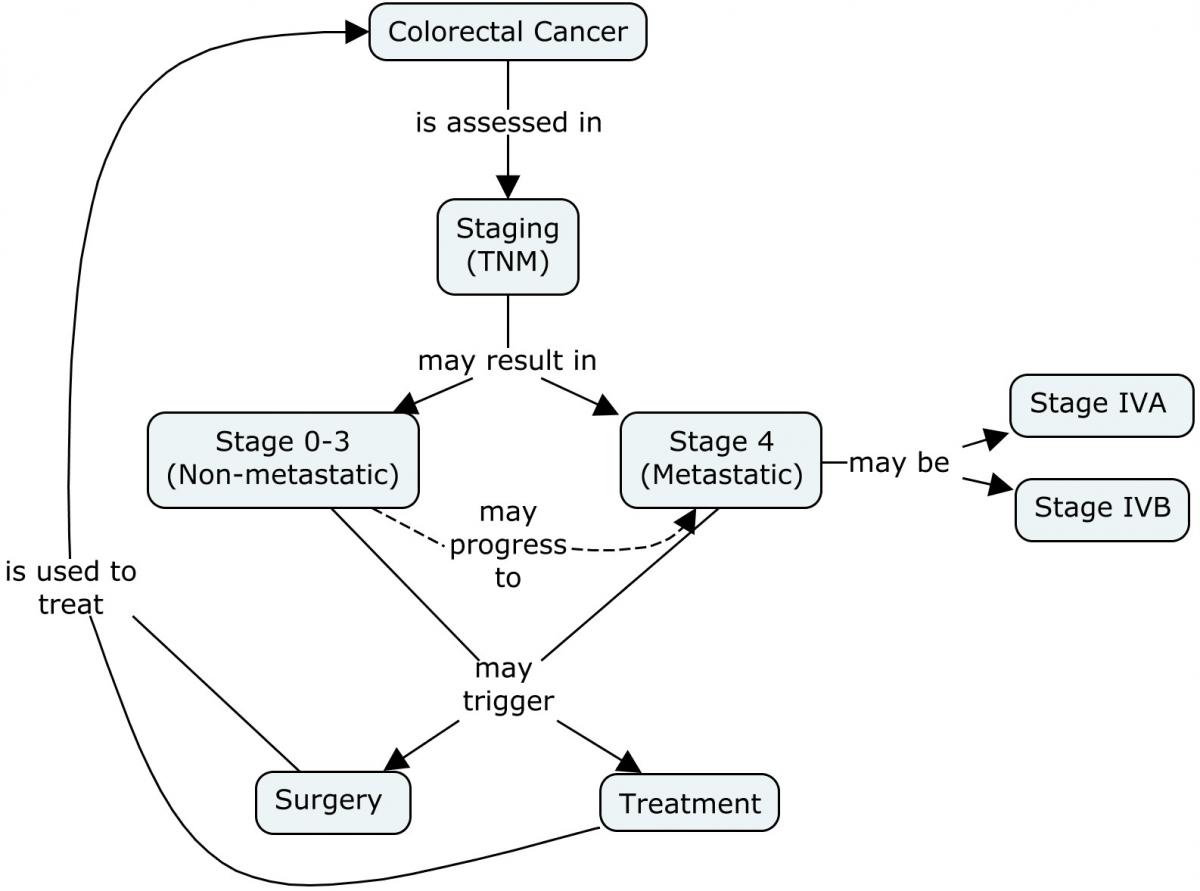
3 Subject and Disease Characteristics
This section provides details of subject and disease characteristics relevant to colorectal cancer.
3.1 Initial Diagnosis
Colon cancer is cancer of the large intestine and rectal cancer is cancer of the last several inches of the colon. Together, they are often referred to as colorectal cancers. Most colorectal cancer begins as a growth of a polyp (in particular, an adenomatous polyp), which may form on the inner wall of the colon or rectum. Screening and work-up for colorectal cancer may include:
- Fecal occult blood test, DNA stool tests (note that DNA stool tests are still exploratory in Japan and not commonly performed)
- Flexible sigmoidoscopy, barium enema, or colonoscopy
- Clinical examinations (e.g., digital rectal examination)
- A biopsy or polypectomy, performed during a test to determine whether cancer is present
- Imaging such as magnetic resonance imaging (MRI), computed tomography (CT), positron emission tomography/computed tomography (PET/CT), ultrasound (US), bone scan, virtual colonoscopy
- Laboratory testing and molecular work-up (including serum carcinoembryonic antigen [CEA] and DNA testing)
For clinical trials, data collected on initial diagnosis of colorectal cancer include the age at initial diagnosis, the date of diagnosis, family history, and diagnostic work-up (including location of primary tumor, histology, and molecular work-up). Studies in metastatic colorectal cancer may collect data both on the initial diagnosis and from additional diagnostic work-ups (including molecular work-up) conducted when the subject enters the study. Several genes have been identified as being associated with colorectal cancer.
For the purposes of this TAUG, the definition of diagnosis can refer to the following categories:
- Confirmation that colorectal cancer is present
- Determination of the severity/extent of the colorectal cancer
Concept Map. Initial Diagnosis of Colorectal Cancer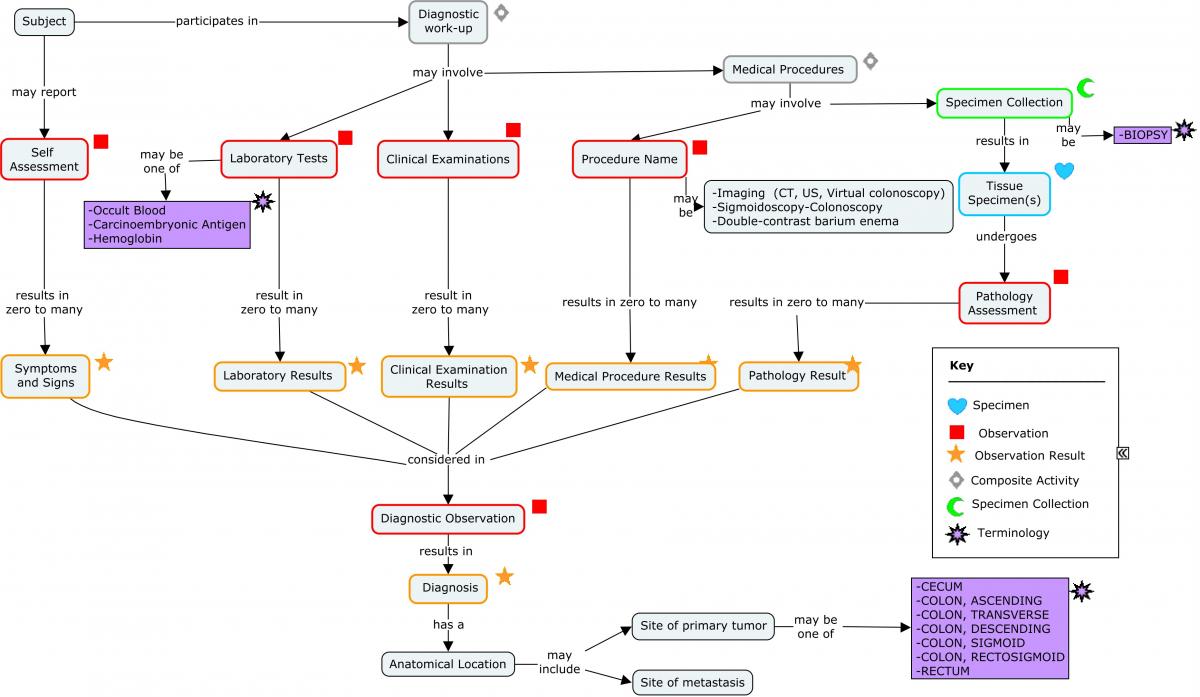
3.2 Staging
Disease staging describes the extent to which the malignancy has spread in the body. It contributes to the determination of treatment options and to the estimation of a patient's prognosis.
The AJCC's Cancer Staging Manual (available at https://cancerstaging.org/Pages/default.aspx) is a named and copyrighted instrument which describes TNM Classification. Guidance on mapping its components to SDTM will be provided as part of a supplement developed by the Questionnaires, Ratings, and Scales (QRS) Team, once permission to do so is granted by the copyright holder. It is anticipated that TNM staging will be represented in the Disease Response and Clin Classification (RS) domain.
T, N, and M stand for primary Tumor, regional lymph Node, and distant Metastasis, respectively.
T, N, and M determinations can be based on indirect measurements from physical examination and imaging tests (clinical staging), and/or on observations made directly on surgically sampled tissue (pathologic staging).
3.3 Pathology
Fo llowing a biopsy, the resected tissue is examined by a pathologist. Pathologic assessments are both macroscopic and microscopic, and provide further detail as to the specific type, staging, and aggressiveness of the cancer. Specimens are prepared and processed as illustrated in the following concept map.
Concept Map. Molecular Work-up: Specimen Collection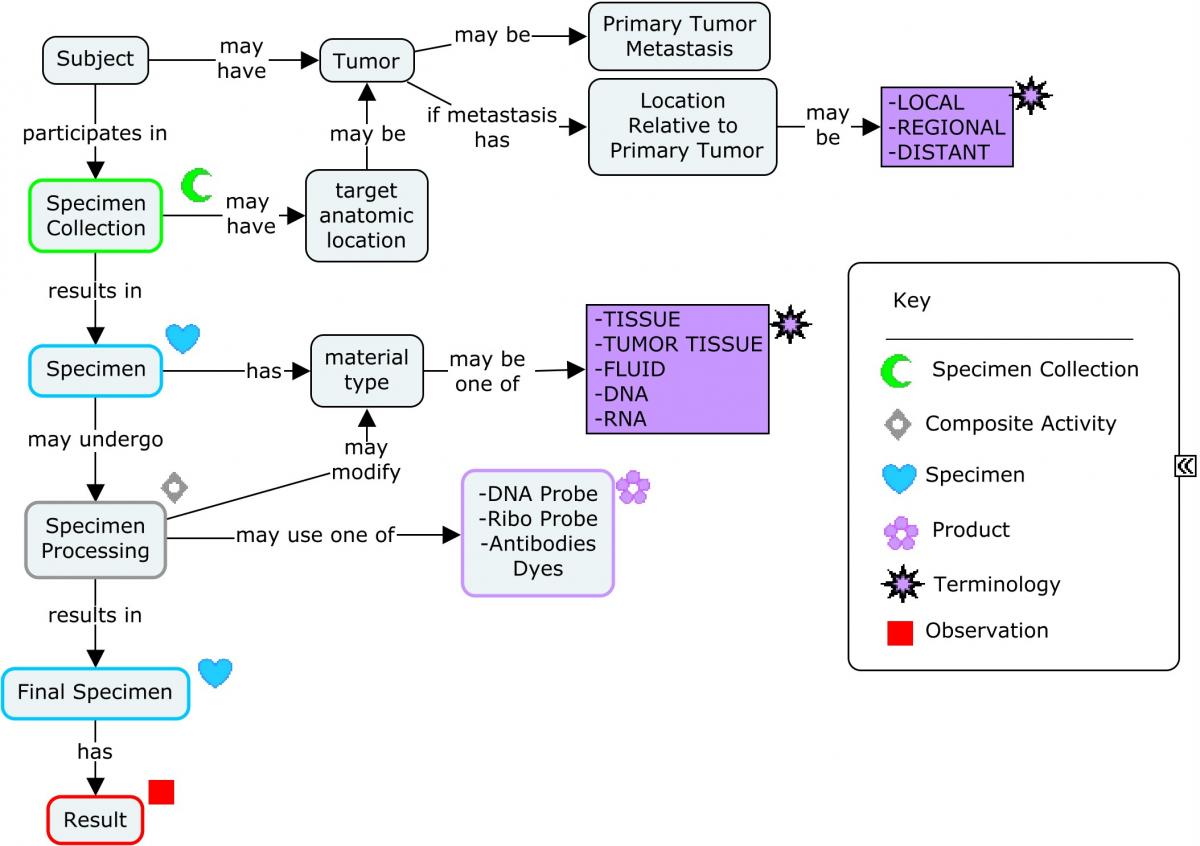
The assessment of genes and proteins helps in the determination of the specific type and aggressiveness of the cancer. The following concept map provides a high-level overview of a molecular work-up that may be performed.
Concept Map. Molecular Work-up: Overview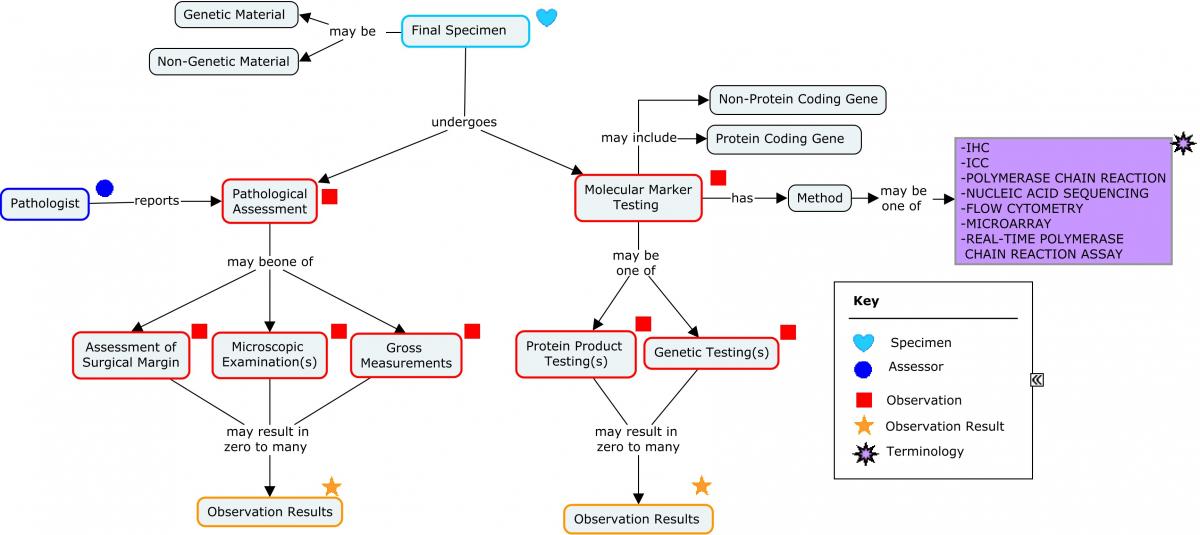
The concept map below may be used to resolve questions about where particular tests should be represented in SDTM domains.
Concept Map. Molecular Work-up: Recommended SDTM Domains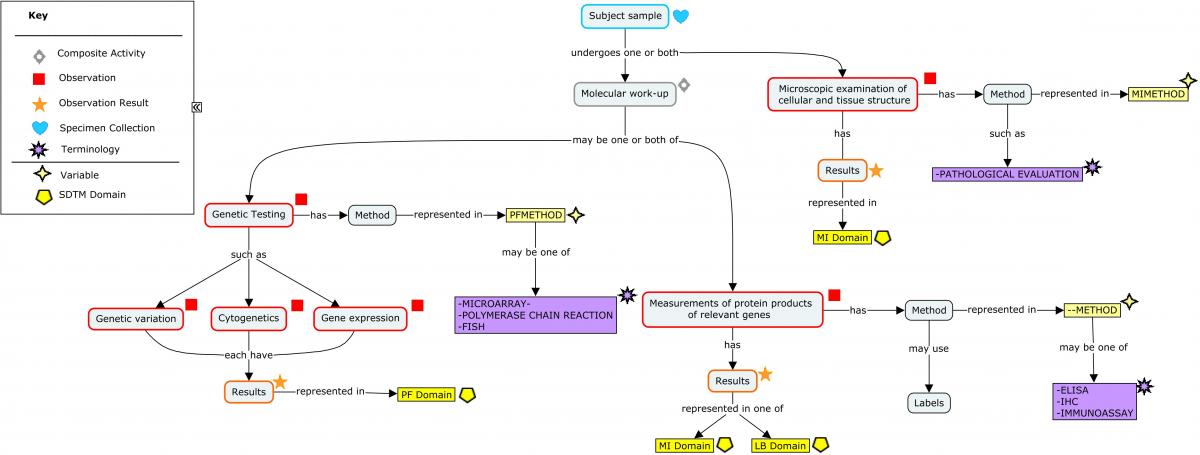
3.3.1 Common Tests/Findings Associated with Colorectal Cancer
The following tables provide details on the more common tests and findings associated with colorectal cancer. The abbreviations and test/finding names in these tables are commonly used in clinical practice. Abbreviations are not necessarily the --TESTCD and --TEST values in CDISC Controlled Terminology. Similarly, the descriptions of these tests and findings are given in the context of colorectal cancer, and may not be Controlled Terminology definitions. When constructing standard datasets, consult the current version of CDISC Controlled Terminology (available at http://www.cancer.gov/cancertopics/cancerlibrary/terminologyresources/cdisc) for values of --TEST and --TESTCD.
3.3.1.1 Prespecified Findings
The Prespecified Findings table below lists findings that may be collected as results of a general microscopic examination looking for abnormalities, or collected via individual questions with present/absent responses. In some cases (e.g., bowel obstruction/perforation) the data may be represented as medical history. Please note that this table is not an exhaustive list but details the more common findings associated with colorectal cancer.
| Common Name (Abbreviation) | Description |
|---|---|
| Lymphovascular Invasion | A microscopic finding that indicates the infiltration of the lymphatic vasculature by a malignant cell population and has been shown to be a stage-independent prognostic indicator for metastatic disease in colorectal cancer.[3] |
| Bowel Obstruction | Partial or complete blockage of the lumen of the bowel, preventing normal flow of the intestinal contents within the bowel. Colorectal cancer is an etiologic agent of bowel obstruction, which has been shown to be associated with poor long-term prognosis in colorectal carcinoma.[4] |
| Bowel Perforation | A rupture in the wall of the small or large intestine due to traumatic or pathologic processes. Colorectal carcinomas can perforate the bowel at the site of disease or cause diastatic perforation at non-involved bowel locations. Perforation at the site of colorectal carcinomas has been shown to be a negative prognostic indicator for 5-year survival.[5] |
3.3.1.2 Laboratory and Microscopic Assessments
The findings in the following table may be collected as results of laboratory tests or microscopic examinations, or via prespecified questions with present/absent responses. Please note that the table does not provide an exhaustive list but rather describes the most common types of colorectal cancer assessments.
| Common Name (Abbreviation) | Description |
|---|---|
| Carcinoembryonic Antigen (CEA) | A cancer-specific antigen associated with both tumors and the developing fetus. Production of the antigen normally ceases shortly before birth, but may reappear in people who develop certain types of cancer, especially colorectal cancers. CEA is a well-characterized non-specific tumor marker that has been shown to correlate with tumor progression and recurrence. Further, preoperative elevated levels of CEA have been shown to be a negative prognostic indicator for overall survival in colorectal cancer.[6] |
| Cancer Antigen 19-9 (CA19-9) | A fucosylated glycosphingolipid carbohydrate antigen that is soluble and adsorbed to erythrocytes and many adenocarcinomas of the digestive tract. CA19-9 is structurally related to the Lewis blood group antigens and is a highly used tumor marker for colorectal cancer. High pre-operative levels of CA19-9 have been shown to have poor prognostic significance and may predict peritoneal recurrence of disease.[7] |
| Lactate Dehydrogenase (LDH) | A family of homotetrameric cytoplasmic enzymes involved in the conversion of L-lactate and NAD to pyruvate and NADH in the final step of anaerobic glycolysis. High serum LDH levels have been variably linked with poor overall survival; LDH levels may also predict response to certain chemotherapies.[8,9] |
| Homeobox Protein CDX-2 (CDX-2) | A 34 kDA homeobox protein encoded by the human CDX2 gene. CDX-2 is a transcription factor that is involved in intestinal morphogenesis and is expressed in most colorectal carcinomas. Lack of CDX2 expression has been shown to have poor prognostic significance for 5-year survival. Individuals with high-risk Stage II colon cancer and lack of CDX2 expression appear to benefit from adjuvant chemotherapy.[10] Please note that this concept is related to the adjuvant setting of colorectal cancer. |
| Mismatch Repair (MMR) | Microsatellite instability results from the inability of the MMR proteins to fix a DNA replication error. It is becoming the standard of care at many centers that all individuals with newly diagnosed CRC are evaluated for Lynch syndrome through molecular diagnostic tumor testing assessing MMR deficiency.[11] |
| Human Epidermal Growth Factor Receptor (HER-2, HER2/neu) | A 138 kDA receptor tyrosine-protein kinase encoded by the human ERBB2 gene. HER2/neu is involved in cell proliferation, tyrosine phosphorylation, and signal transduction. Cytoplasmic and membranous HER-2 over-expression as detected by immunohistochemistry has been variably reported in 15% to 60% of colorectal tumors. Although data are limited and very recent, there is some evidence that targeting HER-2 in metastatic colorectal carcinomas has increased the overall survival in a small number of patients.[12,13] NOTE: The PGx Team has not yet determined in which domain FISH analysis belongs. |
Example
This is an example of how laboratory results can be represented in SDTM for Carcinoembryonic Antigen (CEA) and Cancer Antigen 19-9.
| Row 1: | Shows the results for the assessment of CEA in blood at screening. |
|---|---|
| Row 2: | Shows the results for Cancer Antigen 19-9 in blood at screening. |
lb.xpt
| Row | STUDYID | DOMAIN | USUBJID | LBSEQ | LBTESTCD | LBTEST | LBCAT | LBSPEC | LBMETHOD | LBORRES | LBORRESU | LBSTRESC | LBSTRESU | VISITNUM | VISIT | LBDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | LB | 001-001 | 1 | CEA | Carcinoembryonic Antigen | CHEMISTRY | BLOOD | IMMUNOASSAY | 7 | ng/mL | 7 | ng/mL | 2 | Week 2 | 2013-02-16 |
| 2 | CRCA001 | LB | 001-001 | 2 | CA19_9AG | Cancer Antigen 19-9 | CHEMISTRY | BLOOD | IMMUNOASSAY | 56 | U/mL | 56 | U/mL | 2 | Week 2 | 2013-02-16 |
Example
This is an example of the CDX-2 test that may be conducted in clinical trials in colorectal cancer. The test was performed on tumor tissue using an IHC method.
mi.xpt
| Row | STUDYID | DOMAIN | USUBJID | MISEQ | MITESTCD | MITEST | MIORRES | MISTRESC | MISPEC | MILOC | MIMETHOD | VISITNUM | VISIT | MIDTC | MIDY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | MI | 1201 | 1 | CDX2AG | CDX2 Antigen | 1+ (<25% positive cells) | 1+ (<25% positive cells) | TISSUE | COLON | IHC | 10 | SCREEN | 2013-02-16 | -5 |
3.3.1.3 Genetic and Molecular Analysis
The following table lists genetic and molecular analysis terms related to colorectal cancer. Tests can be run on different sample sources (e.g., tumor tissue, plasma). Please note that this list is not comprehensive but rather includes the most common analyses associated with colorectal cancer.
| Common Name (Abbreviation) | Description |
|---|---|
| Microsatellite Instability Status (MSI Status) | The condition or state of genomic instability associated with defective DNA mismatch repair in tumors. MSI is assessed by determining the difference in expression of microsatellite sequences within 5 of the Bethesda markers, in tumor versus normal tissue.[14] In colorectal carcinoma, MSI has been associated with the anatomical location of the tumor, poor differentiation, and TNM stage.[15] |
| Kirsten Rat Sarcoma Viral Oncogene Homolog gene (KRAS) | This gene present on chromosome 12p21.1 encodes the k-RAS protein, a GTPase that has a role in cell signaling, proliferation, and apoptosis. Mutation can lead to the production of a constitutively activated k-RAS protein that has been linked to the development of many cancers. The identification of a KRAS-activing mutation in colorectal cancer is critical as this subset of patients has not been shown to benefit from therapies that target the EGF receptor (anti-EGFR therapies).[16] |
| Neuroblastoma RAS (NRAS) | The NRAS Viral Oncogene Homolog Gene present on chromosome 1p13.2 encodes the n-RAS protein, a GTPase that has a role in cell signaling, proliferation, and apoptosis. Mutations in amino acids 12, 13, and 61 lead to the production of a constitutively activated n-RAS protein that has been linked to the development of colorectal cancer and resistance to anti-EGFR therapies.[17] |
| BRAF | The B-Raf Proto-Oncogene, Serine/Threonine Kinase gene present on chromosome 7 encodes the b-RAF protein, a serine/threonine kinase that has a role in regulating the MAP kinase/ERK signaling pathway to regulate cell growth and inhibit apoptosis. In colorectal cancer, the V600E mutation leads to a more aggressive phenotype and resistance to anti-EGFR therapies.[18] |
| UDP Glucuronosyltransferase Family 1 Member A1 gene (UGT1A1) | This gene present on chromosome 2 encodes the UDP-glucuronosyltransferase 1-1 protein, a uridine diphosphate glucuronosyltransferase that has a role in the glucuronidation of bilirubin, simple phenols, flavones, and C18 steroids. UGT1A1 polymorphisms have been shown to be associated with irinotecan-induced toxicities in colorectal cancer patients.[19] |
| Adenomatous Polyposis Coli (APC) | The APC gene present on the long arm of chromosome 5 encodes the APC protein, a tumor suppressor that interacts with E-cadherin and controls levels of beta-catenin to regulate cell adhesion. This protein is also involved in inhibition of the Wnt signaling pathway. Various germline mutations in the APC gene, especially within amino acids 1286-1513 or 1061-1309, inactivate the APC protein leading to constitutive activation of the Wnt signaling pathway leading to tumorigenesis. APC mutation is a hallmark of Familial Adenomatous Polyposis, an autosomal dominant disorder characterized by the presence of multiple adenomas in the colon and rectum.[20] |
Examples of how a sponsor represented results of molecular and genetic assessments that may be performed in colorectal clinical trials are provided in the following sections.
The College of American Pathologists "Template for Reporting Results of Biomarker Testing of Specimens From Patients With Carcinoma of the Colon and Rectum" (available at https://www.ncbi.nlm.nih.gov/pubmed/23808403) was referenced when creating these examples.
The Study Data Tabulation Model Implementation Guide for Pharmacogenomics/Genetics future version was considered when the following Pharmacogenomics/Genetics Findings (PF) domain example were created, especially proposed controlled terminology. However, all variables used in these examples were defined in SDTMIG-PGx v1.0. Please note that the variables PFGENRI, PFGENTYP, PFGENLI, PFGENTRG, PFGENLOC, PFGENSR, and PFMUTYP were not standard variables in the current SDTM model at the time this guide was published.
3.3.1.3.1 Microsatellite Instability (MSI) and Mismatch Repair (MMR)
Microsatellite Instability (MSI) and Mismatch Repair (MMR) data collected in a colorectal cancer clinical trial. The Revised Bethesda Guidelines for Hereditary Nonpolyposis Colorectal Cancer (Lynch Syndrome) and Microsatellite Instability(available at https://www.ncbi.nlm.nih.gov/pubmed/14970275) provides guidance on testing that may be performed in colorectal cancer patients. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) recommends testing of colorectal tumors of individuals with newly-diagnosed colorectal cancer.[21]
In the examples below, the investigator was requested to report any information available on MSI or MMR testing performed at the time of the initial diagnosis. MMR IHC testing is used to detect the presence or absence of the protein products of the mismatch repair genes (e.g., MLH1, MSH2, PMS2 and MSH6), whereas MSI PCR testing will determine the DNA mismatch repair loss in tumors.
The SDTM domain used to represent the results depends on the method employed. DNA-based MSI testing was represented in the PF domain, and results for protein-based MMR using IHC methodology were represented in the MI domain.
DNA MSI testing results may be reported as:
- the number of markers exhibiting instability;
- the percentage of markers tested exhibiting instability;
- the specific marker present; or
- an interpretation of the MSI testing, where results are commonly reported using the terminology of MSI-H, MSI-L, and MSS.
Different panels of markers may be tested depending upon the laboratory and kit used.
Example
Sponsors may collect information on the kit used for testing and the characteristics of the kit. If collected, each kit may be treated as a device. The SDTM device domains may then be used to represent the relevant information as described in the Study Data Tabulation Model Implementation Guide for Medical Devices (SDTMIG-MD). The following section describes how this data may be represented.
The Device Identification (DI) domain is used to uniquely identify each specific kit used in the study. This domain provides a consistent sponsor-defined variable (SPDEVID) for linking data across domains. At a minimum, Device Type (DIPARMCD="DEVTYPE") should be recorded. When DIPARMCD is DEVTYPE, the Device Type values stored in DIVAL are subject to terminology defined by the Global Medical Device Nomenclature (GMDN), available at https://www.gmdnagency.org. Refer to the SDTMIG-MD for guidance on terminology that should be used for DIVAL values when DIPARMCD is DEVTYPE.
In the example DI domain below, three different kits were used for MSI testing during the study and the sponsor created a device SPDEVID for each kit.
Note that the DI domain does not include the USUBJID variable because the domain represents information on the device itself and the information is not related to any subject.
| Rows 1-3: | Each kit used for MSI testing was assigned a sponsor-defined SPDEVID. In order to uniquely identify this kit, rows for DIPARMCD and DIPARM of Device Type, Model Number (which was the version of the kit) and Manufacturer were provided. |
|---|---|
| Rows 4-7: | The other kits used for MSI were also assigned a sponsor-defined SPDEVID. In this case, only Device Type and Manufacturer were required to uniquely identify these kits. |
di.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DISEQ | DIPARMCD | DIPARM | DIVAL |
|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | DI | MSI-Kit-1 | 1 | DEVTYPE | Device Type | MSI Analysis Kit |
| 2 | CRCA001 | DI | MSI-Kit-1 | 2 | MODEL | Model Number | Ver 1.2 |
| 3 | CRCA001 | DI | MSI-Kit-1 | 3 | MANUF | Manufacturer | Company X |
| 4 | CRCA001 | DI | MSI-Kit-2 | 1 | DEVTYPE | Device Type | MSI Analysis Kit |
| 5 | CRCA001 | DI | MSI-Kit-2 | 2 | MANUF | Manufacturer | Company Y |
| 6 | CRCA001 | DI | MSI-Kit-3 | 1 | DEVTYPE | Device Type | MSI Analysis Kit |
| 7 | CRCA001 | DI | MSI-Kit-3 | 2 | MANUF | Manufacturer | Company Z |
The Device Properties (DO) domain is used to represent important characteristics of a device that do not form part of the unique sponsor-defined identification of the device provided in DI. These characteristics are properties of the device and can not be changed for each subject. Note that this domain does not include the USUBJID variable because the domain represents information on the device itself and the information is not related to the subject.
In the DO example below, the sponsor reported the marker names tested in the various kits that were used.
| Rows 1-2: | Show the specific mononucleotide and pentanucleotide markers tested by MSI-Kit-1. |
|---|---|
| Row 3: | Shows the specific mononucleotide tested by MSI-Kit-2. This kit only tests m ononucleotide markers. |
| Rows 4-5: | Show the specific mononucleotide and dinucleotide markers tested by MSI-Kit-3. |
do.xpt
| Row | STUDYID | DOMAIN | SPDEVID | DOSEQ | DOTESTCD | DOTEST | DOORRES |
|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | DO | MSI-Kit-1 | 1 | MONTMKNM | Mononucleotide Marker Names | BAT-25, BAT-26, MONO-27, NR-21, NR-24 |
| 2 | CRCA001 | DO | MSI-Kit-1 | 2 | PENTMKNM | Pentanucleotide Marker Names | Penta C, Penta D |
| 3 | CRCA001 | DO | MSI-Kit-2 | 1 | MONTMKNM | Mononucleotide Marker Names | BAT-25, BAT-26, MONO-27, NR-21, NR-24 |
| 4 | CRCA001 | DO | MSI-Kit-3 | 1 | MONTMKNM | Mononucleotide Marker Names | BAT-25, BAT-26 |
| 5 | CRCA001 | DO | MSI-Kit-3 | 2 | DINTMKNM | Dinucleotide Marker Names | D2S123, D5S346, D17S250 |
Since no subject-specific set-up was required, therefore the Device In-Use domain (DU) is not shown.
The Device-Subject Relationships (DR) domain is used to link each subject to the associated device. Information on only 4 subjects is provided as an example. This domain should be included when devices of interest are under study. When the devices are not under study, this domain may not be needed. If it is important to know the individual device used with each subject, this domain should be included. In this case, the sponsor included the DR domain.
dr.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID |
|---|---|---|---|---|
| 1 | CRCA001 | DR | 1001 | MSI-Kit-2 |
| 2 | CRCA001 | DR | 1002 | MSI-Kit-1 |
| 3 | CRCA001 | DR | 1003 | MSI-Kit-2 |
| 4 | CRCA001 | DR | 1004 | MSI-Kit-3 |
The MSI test results are illustrated below for MSI-summary results and MSI-individual marker results. It is recommended that users discuss with the relevant regulatory authority whether to represent summary results and individual markers. Both summary results and individual marker results can be presented in one dataset. However, in the examples that follow the datasets were split to enhance the readability for users. When datasets are combined, sponsors may use PFGRPID to link the appropriate records together.
Example
In this example of MSI summary results, the sponsor collected information on the kit used for testing. The variable SPDEVID is included to link the kit used for testing to the Device domains that provide the kit characteristics. Many sponsors may not collect information on the kit used for testing. Sponsors typically may collect only one of the tests illustrated below. Because these are summary results across several markers, the expected variable PFGENRI is included in the dataset but is null. However, the specific markers tested were described in the device domains as previously illustrated.
| Row 1: | Shows the number of markers exhibiting instability for Subject 1001. |
|---|---|
| Row 2: | Shows the percentage of markers exhibiting instability for Subject 1001. |
| Rows 3-5: | Show the overall status of the DNA MSI testing for three subjects. The results are reported as MSI-H, MSI-L, MSH-S, or MSI-Indeterminate. The result of MSI-Indeterminate indicates that testing was performed, but a specific status could not be determined. |
| Row 6: | Shows that the DNA MSI testing was not performed (Not Done) for Subject 1004. |
pf.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | PFSEQ | PFTESTCD | PFTEST | PFGENRI | PFGENTYP | PFCAT | PFORRES | PFORRESU | PFSTRESC | PFSTRESN | PFSTRESU | PFSTAT | PFSPEC | PFMETHOD | VISITNUM | VISIT | PFDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | PF | 1001 | MSI-Kit-2 | 1 | NUMMISTB | Num Nuc Markers Exhibit Instability | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | 4 | 4 | 4 | DNA | PCR | 10 | BASELINE | 2014-10-10 | ||||
| 2 | CRCA001 | PF | 1001 | MSI-Kit-2 | 2 | PCTMISTB | Pct Nuc Markers Exhibit Instability | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | 80 | % | 80 | 80 | % | DNA | PCR | 10 | BASELINE | 2014-10-10 | ||
| 3 | CRCA001 | PF | 1001 | MSI-Kit-2 | 3 | MSIOSTAT | Microsatellite Instability Overall Stat | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | MSI-H | MSI-H | DNA | PCR | 10 | BASELINE | 2014-10-10 | |||||
| 4 | CRCA001 | PF | 1002 | MSI-Kit-1 | 1 | MSIOSTAT | Microsatellite Instability Overall Stat | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | MSI-L | MSI-L | DNA | PCR | 10 | BASELINE | 2014-09-10 | |||||
| 5 | CRCA001 | PF | 1003 | MSI-Kit-2 | 1 | MSIOSTAT | Microsatellite Instability Overall Stat | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | MSI-INDETERMINATE | MSI-INDETERMINATE | DNA | PCR | 10 | BASELINE | 2015-12-10 | |||||
| 6 | CRCA001 | PF | 1004 | 1 | MSIOSTAT | Microsatellite Instability Overall Stat | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | NOT DONE | 10 | BASELINE | 2015-02-13 |
Example
This example of MSI individual results illustrates how the results for each specific MSI nucleotide marker were represented. The information on the kit used for testing was collected by the sponsor. The variable SPDEVID is included to link the kit used for testing to the Device domains that provide the kit characteristics. Sponsors may decide to derive the summary MSI results in ADaM if reporting results for each nucleotide marker. Each row represents a marker that was included in the MSI testing. In this situation, the nucleotide marker name is included in PFGENRI. The PFTEST results indicate whether the marker was Stable or Unstable. SPDEVID links the subject to the MSI kit used for each subject.
pf.xpt
| Row | STUDYID | DOMAIN | USUBJID | SPDEVID | PFSEQ | PFGRPID | PFTESTCD | PFTEST | PFGENRI | PFGENTYP | PFCAT | PFORRES | PFSTRESC | PFSPEC | PFMUTYP | PFMETHOD | VISITNUM | VISIT | PFDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | PF | 1005 | MSI-Kit-3 | 1 | 1 | MICRISTB | Microsatellite Instability | BAT25 | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | STABLE | STABLE | DNA | GERMLINE | POLYMERASE CHAIN REACTION | 10 | BASELINE | 2014-10-10 |
| 2 | CRCA001 | PF | 1005 | MSI-Kit-3 | 2 | 1 | MICRISTB | Microsatellite Instability | BAT26 | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | STABLE | STABLE | DNA | GERMLINE | POLYMERASE CHAIN REACTION | 10 | BASELINE | 2014-10-10 |
| 3 | CRCA001 | PF | 1005 | MSI-Kit-3 | 3 | 1 | MICRISTB | Microsatellite Instability | D2S123 | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | UNSTABLE | UNSTABLE | DNA | GERMLINE | POLYMERASE CHAIN REACTION | 10 | BASELINE | 2014-10-10 |
| 4 | CRCA001 | PF | 1005 | MSI-Kit-3 | 4 | 1 | MICRISTB | Microsatellite Instability | D5S346 | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | STABLE | STABLE | DNA | GERMLINE | POLYMERASE CHAIN REACTION | 10 | BASELINE | 2014-10-10 |
| 5 | CRCA001 | PF | 1005 | MSI-Kit-3 | 5 | 1 | MICRISTB | Microsatellite Instability | D17S250 | MICROSATELLITE | MOLECULAR DIAGNOSTIC TESTING | STABLE | STABLE | DNA | GERMLINE | POLYMERASE CHAIN REACTION | 10 | BASELINE | 2014-10-10 |
The following modeling of MMR Proteins in SDTM is under discussion and users are cautioned that this modeling may be subject to change.
Example
Immunostaining of the tumor tissue is used to determine the nuclear expression of the MMR Proteins, MLH1, MSH2, MSH6 and PMS2. MMR protein expression is defined as the presence of nuclear staining within the tumor regardless of intensity or the number of positive nuclei.
| Row 1: | Shows the interpretation of the IHC MMR testing. The original results are reported using the categories presented in the College of American Pathologists (CAP) template for reporting results of biomarker testing of specimens from patients with carcinoma of the colon and rectum.[22] Because this is an interpretation of the results for the staining of all the relevant proteins, a specific test was used to represent the result. |
|---|---|
| Rows 2-5: | Each row represents the expression of the MMR proteins in the tumor cells. The protein of interest is provided in the MITESTCD/MITEST. |
mi.xpt
| Row | STUDYID | DOMAIN | USUBJID | MISEQ | MITESTCD | MITEST | MIORRES | MISTRESC | MISPEC | MILOC | MIMETHOD | VISITNUM | VISIT | MIDTC | MISCELOC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | MI | 1002 | 1 | MMRPINTP | Mismatch Repair Proteins Interpretation | NO LOSS OF NUCLEAR EXPRESSION OF MMR PROTEINS | NO LOSS OF NUCLEAR EXPRESSION OF MMR PROTEINS | TUMOR TISSUE | COLON | IHC | 10 | BASELINE | 2014-09-10 | NUCLEUS | |
| 2 | CRCA001 | MI | 1002 | 2 | MLH1 | MutL Homolog 1 | INTACT NUCLEAR EXPRESSION | INTACT NUCLEAR EXPRESSION | TUMOR TISSUE | COLON | IHC | 10 | BASELINE | 2014-09-10 | NUCLEUS | |
| 3 | CRCA001 | MI | 1002 | 3 | MSH2 | MutS Homolog 2 | LOSS OF NUCLEAR EXPRESSION | LOSS OF NUCLEAR EXPRESSION | TUMOR TISSUE | COLON | IHC | 10 | BASELINE | 2014-09-10 | NUCLEUS | |
| 4 | CRCA001 | MI | 1002 | 4 | MSH6 | MutS Homolog 6 | INTACT NUCLEAR EXPRESSION | INTACT NUCLEAR EXPRESSION | TUMOR TISSUE | COLON | IHC | 10 | BASELINE | 2014-09-10 | NUCLEUS | |
| 5 | CRCA001 | MI | 1002 | 5 | PMS2 | PMS1 Homolog 2 | UNDETERMINED | UNDETERMINED | TUMOR TISSUE | COLON | IHC | 10 | BASELINE | 2014-09-10 | NUCLEUS |
MI NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| MISCELOC | Subcellular Location | text | Non-Standard Record Qualifier | CRF |
3.3.1.3.2 Genetic Variation
In this example colorectal cancer trial, the sponsor planned exploratory analyses involving tumor-associated somatic variations (mutations), so they collected any available data on genetic variations included in the subject's historical records (e.g., KRAS, NRAS, BRAF, PIK3CA). The amount of available information varied across subjects. Some subjects only had information on whether any variations were detected for each gene (or protein) of interest, whereas other subjects had information on the actual variation detected in each gene (or protein) of interest. The method may or may not have been available. Because the name of the vendor was collected, the sponsor may have the ability to obtain more technical information about the specific tests. These technical details were not modeled in any SDTM dataset.
The sponsor carefully considered the limitations of historical data collection reported by investigators on CRFs and represented this data based on a pragmatic approach. When collecting pre-study data on CRFs, certain data (e.g., reference sequence) may not be readily available. If data are collected using a central vendor, the sponsor should work with the central vendor to ensure that the required details are provided. This TAUG does not include an example of data collected using a central vendor. However, this sponsor collected the name of the vendor used for testing, and this allowed the sponsor to obtain more technical information about the specific tests (if needed for analysis).
This example deviates from the current recommendation in the SDTMIG-PGx v1.0 as the information on the genetic variations were not parsed out using the variables PFORRES, PFORREF, and PFGENLOC. This parsing out is recommended, but in this example the sponsor did not want to manually parse out the information to avoid potential errors. This pragmatic representation of the genetic data is under discussion within the PGx Team. Sponsors are urged to discuss such pragmatic representation with the appropriate regulatory agency. For the reader's information, the appropriate parsing of "c.1637A>T" (indicates nucleotide 1637 an A is changed to a T) is: PFORREF would be A, the PFORRES would be T, and PFGENLOC would be 1637.
However, if a central lab was used, the PGx Team recommends that the genetic data be parsed out into the appropriate items.
In addition, the published controlled terminology for PFTEST "Genetic Variation" was used instead of the PFTEST values of "Amino Acid" or "Nucleotide" that are suggested in the SDTMIG-PGx v1.0.
This example also used the CAP reporting template (available at https://www.ncbi.nlm.nih.gov/pubmed/23808403) as a possible CRF collection format.
Sponsors may collect information on the kit used for testing and the characteristics of the kit. If collected, each kit may be treated as a device. The SDTM device domains may be used to represent the relevant information. See Section 3.3.1.3.1, Microsatellite Instability (MSI) and Mismatch Repair (MMR), for examples on how to show information on a kit used for testing.
When looking at the example below please bear in mind the following:
- The representation of the data in the PF domain focused on ensuring consistency of the information within the dataset. The sponsor decided to include the results of the question whether or not a generic variation was detected in the SDTM-based dataset. Some sponsors may decide that when a variation is detected, the question indicating that the variation was detected may not be represented in the SDTM-based dataset as the specific variation are represented.
- The SDTM example reports the results based on the actual data collected. When CRFs are used to collect pre-study results, sponsors may allow various formats for reporting the results to accommodate the various formats that may have been used when the original data was reported by the lab.
- The protein variation may use the single-letter amino acid symbols or the 3-letter amino acid symbols. For example, a variation in a protein in which the 12th amino acid is Glycine in the reference sequence and Alanine in the subject's sequence can be represented as either Gly12Ala or G12A. Both 3-letter and single-letter symbols for amino acids were published by the International Union of Pure and Applied Chemistry (IUPAC) and the International Union of Biochemistry (IUB) in their 1983 recommendations for the nomenclature and symbolism for amino acids and peptides (available at http://iupac.org/publications/pac/56/5/0595/). Many labs report the results using either the 3-letter or single-letter amino acid names. The CAP reporting template (available at https://www.ncbi.nlm.nih.gov/pubmed/23808403) uses the 3-letter abbreviation for reporting some the amino acid (protein) names. Because the 3-letter amino acid symbols were used, they are reported in the SDTM variable PFORRES/PFSTRESC. This avoided the manual translation of the data into another format. A sponsor is encouraged to design the data collection instrument using the most appropriate format that avoids any manual translation of the data. Sponsors should provide instructions to the site regarding the nomenclature to use within a specific clinical trial. The Human Genome Variation Society (HGVS) nomenclature may be used to report information regarding DNA, RNA, and protein sequences; these standards were updated in 2017.[23]
- A list of pre-specified gene variations may be defined on a CRF either because a set of targeted tests was performed for the study in order to identify specific variations, or as a data entry convenience when collecting information about tests that may have been performed in the past. If a set of targeted tests was performed for a study then, as described in the SDTMIG-PGx v1.0, the PFGENLI and PFGENTRG variables should be used to define the pre-specified variations, PFORRES should be used to indicate whether the variation was detected, and any detected variations should be recorded in PFSTRESC using standard nomenclature. However, when using a pre-specified list of gene variations as a data entry convenience to collect information about tests that may have been performed in the past, sponsors may choose either to transcribe previous results to the format described above or to collect the results in their originally reported format. In this example study, the sponsor chose to represent any variations selected from the pre-specified list or written in by the investigator using the original collection format. PFSTRESC, the standardized result, also used the format associated with the collected variation to avoid any manual translation to another format (e.g., p.Gly12Asp to c.35G>A). Note that the PFSTREC still followed the convention of reporting an amino acid using p. and a nucleotide using c.
- Typically for variations that are not pre-specified, PFGENLOC and PFORREF would be populated. PFORREF would be compared with PFORRES to determine if a mutation is present. However, in this case, only subjects with a mutation had the variation reported. Because the data was collected on CRFs, and these fields would have to be manually populated, the sponsor elected not to include these "permissible" variables in the SDTM-based dataset.
- The PFDTC is the date of the original specimen collection. PFDY is based on the date of the original sample, and not the date the testing was performed.
- Some variables (e.g., PFREFSEQ) are not populated. In this study, the sponsor did not consider this information needed to interpret the data.
- PFRUNDTC is a new proposed variable being considered to be added to the PF domain. This variable is used to represent the date that the sample was analyzed. Because this variable has not be approved, it is represented as an NSV variable.
- In this study, the sponsor standardized the results in an ADaM dataset to facilitate data analysis. This ADaM dataset is not shown.
Example
In this example, the tumor specimen used for testing could have been collected at the time of the original diagnosis, during the prior treatment of the subject, or at entry into the study. When the original testing was performed more than a year before the trial entry, the sponsor requested the investigator to obtain an archival sample for re-testing at a local lab. When reporting the results of the testing (see PF domain), the date of the original sample collection is always represented in PFDTC. Because re-testing on an archival sample may have been performed, the sponsor collected information on specimen tracking in the Biospecimen Events (BE) domain. If re-testing was performed, only the re-testing results were provided.
| Rows 1, 3: | Show the date the original sample was collected in BEDTC and the start date (BESTDTC) of the event represented in BETERM for Subjects 3001 and 3006. In this case, the BEDTC and BESTDTC are the same. Because the end date/time of the event is the same as the start date/time for the event, BEENDTC is null. BEENDTC is included in the dataset because it is an expected variable. |
|---|---|
| Rows 2, 4: | Show the information on the archival sample used for testing. The date the original sample was collected is represented in BEDTC and the start date of the archival retrieval event (BETERM="Archival Sample Retrieval") for Subjects 3001 and 3006. |
| Row 5: | Shows the date that the original sample was collected. No archival samples were processed. |
be.xpt
| Row | STUDYID | DOMAIN | USUBJID | BESEQ | BETERM | BEDECOD | VISITNUM | VISIT | BEDTC | BESTDTC | BEENDTC |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | BE | 3001 | 1 | Sample Collection | COLLECTING | 1 | BASELINE | 2007-05-05 | 2007-05-05 | |
| 2 | CRCA001 | BE | 3001 | 2 | Archival Sample Retrieval | ARCHIVE RETRIEVING | 1 | BASELINE | 2007-05-05 | 2015-06-05 | |
| 3 | CRCA001 | BE | 3006 | 1 | Sample Collection | COLLECTING | 1 | BASELINE | 2009-06-10 | 2009-06-10 | |
| 4 | CRCA001 | BE | 3006 | 2 | Archival Sample Retrieval | ARCHIVE RETRIEVING | 1 | BASELINE | 2009-06-10 | 2015-04-09 | |
| 5 | CRCA001 | BE | 3008 | 1 | Sample Collection | COLLECTING | 1 | BASELINE | 2015-07-02 | 2015-07-02 |
The information on gene variations, which was collected on a CRF in this study, is illustrated below. The data included information on whether or not a variation was detected in a gene and, if variable in the gene was detected, the variation detected was either selected from a pre-specified list of variations or entered in a specify field. Note that, in order to save space, example results for only five subjects is shown and some expected SDTM variables have been omitted. Any unknown results were reported as "NOT DONE" by the sponsor. These "NOT DONE" data were not illustrated in the example, as this as been adequately described in the SDTMIG.
| Rows 1-2: | Show the subject had KRAS mutation detected, and no PIK3CA mutations detected. The specific KRAS variation was not available. |
|---|---|
| Row 3: | Shows the subject had KRAS mutation detected. |
| Rows 4-5: | Show the 2 detected KRAS mutations, 1 in Codon 12 ("Gly12Asp") and 1 in Codon 13 ("Gly13Ser"), reported as separate rows. PFORRES is populated with the variation using the collected format. T he Codon 12 variation was reported using the pre-specified list of KRAS variations on the CRF, but the Codon 13 variation was reported as a write-in result because it was not present on the pre-specified list. The pre-specified list of variations was used as a data entry convenience for this study, so pre-specified variations and write-in variations are represented in the same way in the data. PFSTRESC, the standardized result, also contains the protein format with the 3-letter amino acid letters because there was no manual translation to another format (e.g., p.Gly12Asp to c.35G>A). |
| Row 6: | For the PIK3CA gene, the CRF (see the CAP template referenced above) collected whether a variation was not detected, or a variation was detected at either Exon 9 or Exon 20. Subject 3006 had a variation reported at Exon 9 and PFGENSR was used to report the location of the variation as Exon 9. |
| Row 7: | If a variation of the PIK3CA gene was detected, the specific variation was collected. The PIK3CA variation (c.1637A>T) was represented in both PFORRES and PFSTRESC. Note that this format was used by the investigator when the variation was reported on the CRF. |
| Row 8: | For the PTEN gene, the CRF (see the CAP template referenced above) collected whether a variation was not detected, or a variation was detected at Exon 1-9. Subject 3009 had a variation detected at Exon 1-9. |
| Row 9: | Subject 3009 had a PTEN variation reported. The location of the variation was collected on the CRF, and represented in PFGENSR. Note the investigator specified the variation (c.477G>T) on the CRF. The format used by the investigator to report this variation was represented in PFORRES and PFSTRESC. |
| Row 10: | Subject 3010 had a BRAF mutation but the detected variation was not available. |
pf.xpt
| Row | STUDYID | DOMAIN | USUBJID | PFSEQ | PFTESTCD | PFTEST | PFGENRI | PFGENTYP | PFCAT | PFORRES | PFORRESU | PFGENSR | PFSTRESC | PFMUTYP | PFNAM | PFMETHOD | VISITNUM | VISIT | PFDTC | PFDY | PFRUNDTC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA001 | PF | 3001 | 1 | GENVAR | Genetic Variation | KRAS | GENE | MOLECULAR DIAGNOSTIC TESTING | DETECTED | DETECTED | SOMATIC | GENLAB1 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2007-05-05 | -2957 | 2015-06-05 | |||
| 2 | CRCA001 | PF | 3001 | 2 | GENVAR | Genetic Variation | PIK3CA | GENE | MOLECULAR DIAGNOSTIC TESTING | NOT DETECTED | NOT DETECTED | SOMATIC | GENLAB1 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2007-05-05 | -2957 | 2015-06-05 | |||
| 3 | CRCA001 | PF | 3006 | 1 | GENVAR | Genetic Variation | KRAS | GENE | MOLECULAR DIAGNOSTIC TESTING | DETECTED | DETECTED | SOMATIC | GENLAB2 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2009-06-10 | -2135 | 2015-04-09 | |||
| 4 | CRCA001 | PF | 3006 | 2 | GENVAR | Genetic Variation | KRAS | GENE | MOLECULAR DIAGNOSTIC TESTING | Gly12Asp | p.Gly12Asp | SOMATIC | GENLAB2 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2009-06-10 | -2135 | 2015-04-09 | |||
| 5 | CRCA001 | PF | 3006 | 3 | GENVAR | Genetic Variation | KRAS | GENE | MOLECULAR DIAGNOSTIC TESTING | Gly13Ser | p.Gly13Ser | SOMATIC | GENLAB2 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2009-06-10 | -2135 | 2015-04-09 | |||
| 6 | CRCA001 | PF | 3008 | 1 | GENVAR | Genetic Variation | PIK3CA | GENE | MOLECULAR DIAGNOSTIC TESTING | DETECTED | EXON 9 | DETECTED | SOMATIC | GENLAB3 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2015-07-02 | -20 | 2015-07-02 | ||
| 7 | CRCA001 | PF | 3008 | 2 | GENVAR | Genetic Variation | PIK3CA | GENE | MOLECULAR DIAGNOSTIC TESTING | c.1637A>T | EXON 9 | c.1637A>T | SOMATIC | GENLAB3 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2015-07-02 | -20 | 2015-07-02 | ||
| 8 | CRCA001 | PF | 3009 | 1 | GENVAR | Genetic Variation | PTEN | GENE | MOLECULAR DIAGNOSTIC TESTING | DETECTED | EXON 1-9 | DETECTED | SOMATIC | GENLAB4 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2015-02-18 | -15 | 2015-02-23 | ||
| 9 | CRCA001 | PF | 3009 | 2 | GENVAR | Genetic Variation | PTEN | GENE | MOLECULAR DIAGNOSTIC TESTING | c.477G>T | EXON 5 | c.477G>T | SOMATIC | GENLAB4 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2015-02-18 | -15 | 2015-02-23 | ||
| 10 | CRCA001 | PF | 3010 | 1 | GENVAR | Genetic Variation | BRAF | GENE | MOLECULAR DIAGNOSTIC TESTING | DETECTED | DETECTED | SOMATIC | GENLAB4 | POLYMERASE CHAIN REACTION | 1 | BASELINE | 2015-07-18 | -23 | 2015-07-25 |
PF NSV Metadata
| Variable | Label | Type | Role | Origin |
|---|---|---|---|---|
| PFRUNDTC | Run Date | datetime | Non-Standard Timing | CRF |
3.3.1.4 Histologic and Gross Assessments
The table below lists gross assessments and histologic findings associated with colorectal cancer, including diagnostic terms derived from conventional light microscopic examination of the tissues. Please note that the table is not an exhaustive list.
| Common Name (Abbreviation) | Description |
|---|---|
| Adenocarcinoma | The most common type of colorectal carcinoma. It is characterized by the presence of malignant glandular epithelial cells invading through the muscularis mucosa into the submucosa. Histologic variants include mucinous adenocarcinoma, signet ring cell carcinoma, medullary carcinoma, serrated adenocarcinoma, cribriform comedo-type adenocarcinoma, and micropapillary adenocarcinoma.[24] Japanese classification of histologic subtypes of colorectal carcinomas may differ. Please refer to the Japanese classification for more information: http://www.jsccr.jp/. |
| Mucinous Adenocarcinoma | An invasive colorectal adenocarcinoma characterized by the presence of extracellular mucin pools that contain malignant glandular epithelial structures. The extracellular mucin pools occupy more than 50% of the malignant lesion.[24] Other histologic subtypes may also be of importance, but are not discussed. Japanese classification of histologic subtypes of colorectal carcinomas may differ. Please refer to the Japanese classification for more information: http://www.jsccr.jp/. |
| Location of Primary Tumor | The anatomic region of the colon or rectum within which the primary tumor originated. In patients with metastatic colorectal cancer, colorectal cancer survival directly correlates with the location of the primary tumor; primary tumors arising in the distal colon (descending and sigmoid colons) correlated with increased 1-year survival post-treatment compared to those primary tumors arising in the proximal colon (cecum, ascending, and transverse colon).[25] |
| Rectal Tumor to Anal Verge Distance | A measurement of the length of the span between the rectal tumor and the anal verge. By definition, rectal tumors are located no more than 15-17 cm from the anal verge;[22] greater distance between the rectal tumor and anal verge may correlate with increased disease free survival.[26,27] Please note that in some geographical regions the anatomical definition of "rectum" may differ, which may affect whether a tumor is classified as a rectal tumor. |
| Site of Metastasis | The anatomic region within which the tumor metastases are found. |
| Number of Metastatic Sites | The total number of anatomic sites that are metastatically involved. |
Example
This example shows representation of a gross pathology result, the distance from a rectal tumor to the anal verge. This morphology result was represented in the Gastrointestinal System Findings (GI) physiology/morphology domain. This is a draft domain, although the domain abbreviation is present in controlled terminology.
gi.xpt
| Row | STUDYID | DOMAIN | USUBJID | GISEQ | GITESTCD | GITEST | GIORRES | GIORRESU | GISTRESC | GISTRESN | GISTRESU | GILOC | GIMETHOD | VISITNUM | VISIT | GIDTC | GIDY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA | GI | 6002 | 1 | RTAVDIS | Rectal Tumor to Anal Verge Distance | 5.1 | cm | 5.1 | 5.1 | cm | RECTUM | ENDOSCOPY | 1 | SCREEN | 2016-04-18 | -100 |
Please note that in some geographical regions the anatomical definition of "rectum" may differ which may affect whether a tumor is classified as a rectal tumor.
4 Disease Assessments
This section provides details of disease assessments relevant to colorectal cancer.
4.1 Treatment
Treatments for colorectal cancer may include elimination of the cancer or controlling the effects of the cancer on the body. Some treatments target only the anatomic region in which the cancer exists; others act on a systemic level. In general, types of treatments are sequential rather than concurrent. The concept map below provides an overview of the treatment options.
Concept Map. Treatment Options Following Colorectal Cancer Diagnosis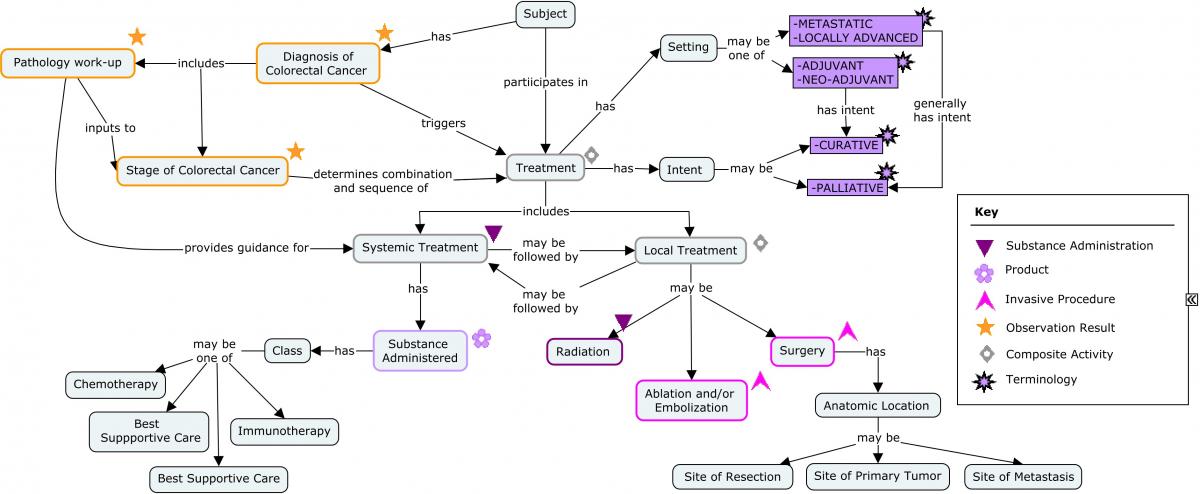
4.1.1 Anti-cancer Therapy
Medications
In cancer trials, it is important to collect information on prior anti-cancer therapies, and any anti-cancer therapies given after the study drug of interest has been discontinued. Typically, in colorectal cancer, anti-cancer treatments are given as regimens.
A regimen is a treatment plan that specifies the dosage, the schedule, and the duration of treatment (NCI definition: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/regimen). In cancer trials, subjects are often prescribed treatment plans that contain multiple treatment components. These multiple treatment plans are commonly known by an acronym (e.g., FOLFOX, FOLFIRI). Sponsors often collect information about the regimen itself (e.g., start date, end date, best response, reason for discontinuation) as well as information on the individual treatment components within the regimen. In other disease areas, subjects are also treated with a combination of treatments but these treatments are often administered as a single product which is a combination of several active ingredients. It is difficult to represent the information about treatments that include multiple components in the SDTM Concomitant Medications (CM) domain.
CDISC is putting together a team that will focus on the SDTM modeling of combination products/regimens. This topic is applicable to many therapeutic areas. CDISC anticipates that this modeling would be published in a separate document providing guidelines for the representation of combination products or combination regimens.
Surgeries
In some colorectal cancer trials, it may be important to collect information on prior anti-cancer surgeries as well as any such surgeries during the study. The type of surgery performed at the time of initial diagnosis depends on the extent of the cancer. In some situations the anus may be weak or damaged, or must be removed, and a colostomy or an ileostomy may be required. Permanent stomas are required when stool cannot pass through its normal route after surgery. Sphincter-sparing surgery is fairly common for some rectal cancers. Depending upon the study, the sponsor may collect the type of surgery performed at the time of the initial diagnosis. Although not shown, this prior surgery information would be represented in the SDTM Procedures (PR) domain.
In some studies, liver resections may be performed for metastases. It is important to provide the dates of surgery and whether disease was found during surgery. Other surgeries with curative intent may also be allowed. If these surgeries are allowed during the study, a sponsor must specify how this will be handled in the analysis of relevant endpoints (e.g., BOR, PFS).
4.2 Disease Assessments and Response
Response Evaluation Criteria In Solid Tumors (RECIST: https://www.ncbi.nlm.nih.gov/pubmed/19097774) is usually used to assess disease response in metastatic colorectal cancer clinical trials. In this document, RECIST is used as a generic term to refer to the various versions of the RECIST criteria. However, sponsors should always specify which RECIST version was used in a study.
The response criteria chosen to assess disease response may depend on the type of therapy being evaluated. Due to the different response patterns in subjects receiving anti-cancer immunotherapies, other criteria such (e.g., irRC, irRECIST, iRECIST) may also be used.
irRC (immune related response criteria)
The irRC criteria were proposed by Wolchok and colleagues[28] to determine the extent of anti-tumor response to an immunotherapeutic agent. The irRC criteria were based on the WHO tumor response criteria (http://apps.who.int/iris/handle/10665/37200)[29] and bidimensional tumor measurements are used.
irRECIST (irRC using unidimensional measurements)
Mizuki Nishino and colleagues[30] have proposed new criteria using 1-dimensional lesion measurements following RECIST. A poster presented by Bohnsack[31] provides a table comparing irRC and irRECIST immunotherapy criteria.
iRECIST (RECIST 1.1 for immune based therapeutics)
A March 2017 publication by the RECIST Working Group Team[32] describes the consensus iRECIST guideline intended to standardize design and collection guidelines for the evaluation of disease in cancer immunotherapy trials.
Other CDISC oncology TAUGs have provided examples of RECIST assessments represented in the SDTM. This TAUG only shows examples of the irRC criteria. The Lung Cancer TAUG will provide examples of the newly published standardized iRECIST criteria. This TAUG does not provide any recommendation regarding which criteria to use in a clinical trial. Sponsors should always consult regulatory authorities to obtain advice on this topic.
The following concept map provides an overview of the measurement of tumor burden and response assessments.
Concept Map. Assessment of Disease Response in Colorectal Cancer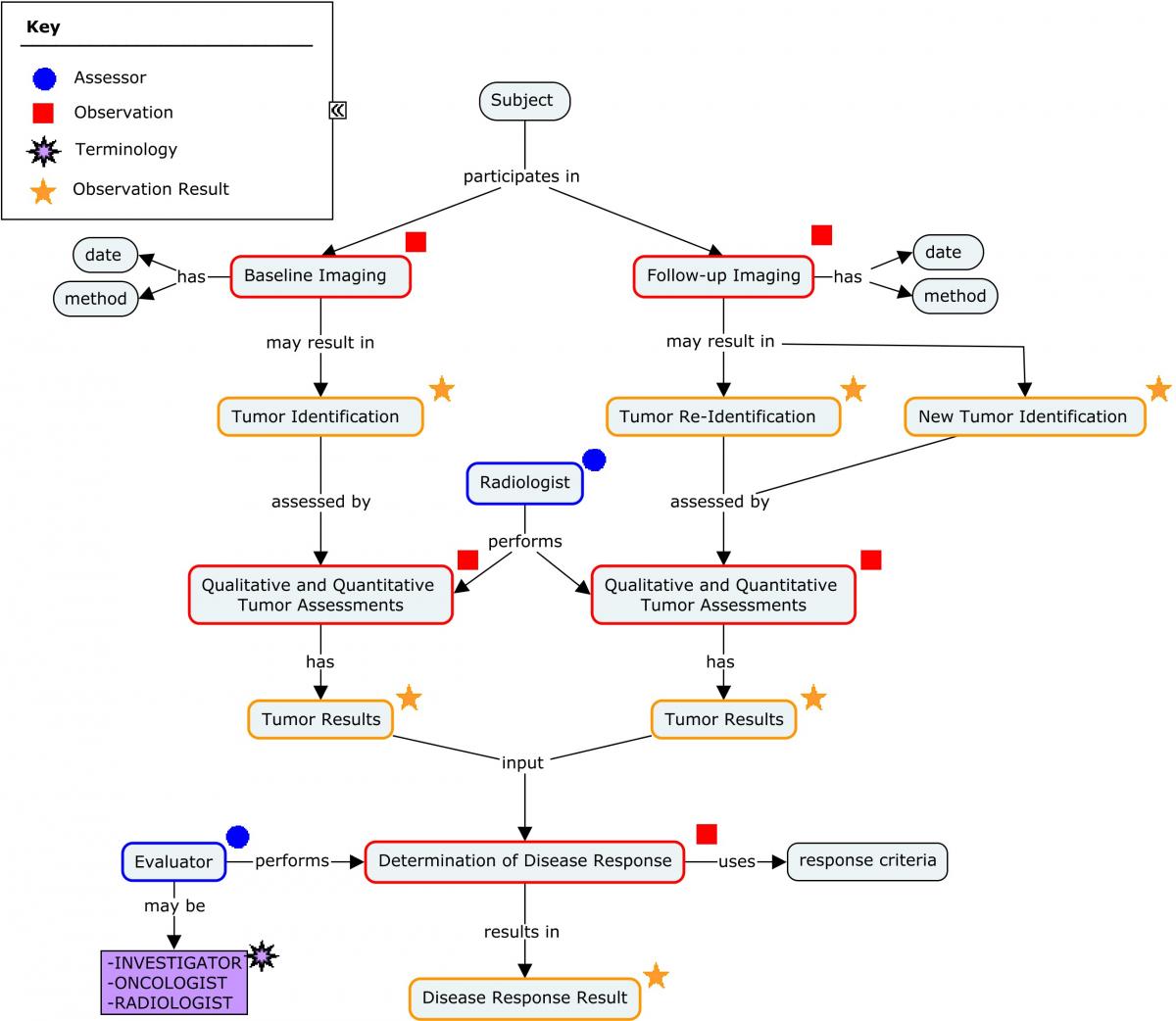
Tumor Results referenced in the previous concept map are based on the response criteria and version used by the sponsor. In RECIST, results are based on the sum of the diameters (longest for non-nodal lesions, short axis for nodal lesions) for all target lesions (Target), with qualitative assessments of non-measurable tumors (Non-Target). The development of new lesions is also assessed. irRC use the WHO criteria for tumor response[29]. In irRC, results are based on the sum of the diameters of bidimensional measurable tumors (Index Lesions + New lesions). Non-measurable lesions are not considered in progression, but are considered in complete response.
It is recommended that the user be familiar with the tumor response guidelines before reviewing the following examples. The sponsor should determined the relevant version of the guideline to be used in a study. General guidance on managing data pertaining to the identification, monitoring, and assessment of tumors and lesions is covered by three SDTMIG Findings domains: Tumor Identification (TU), Tumor Results (TR), and Disease Response (RS). Consult the SDTMIG v3.2 for specifications, assumptions, and examples for these domain models.
The following are examples of tumor identification, tumor assessment, and tumor response for subjects in a colorectal cancer clinical trial. These examples employ the irRC immune-related response criteria. SDTM dataset examples and aCRFs using RECIST have been provided in the TAUGs for Breast Cancer (Section 4.2) and Prostate Cancer (Section 4.2). The sample aCRFs in this TAUG use the irRC criteria.
Annotated CRF: Tumor Identification/Results - irRC - Index Lesions
This CRF is only an example and is not meant to imply that any particular layout or collection plan is preferable over another.
This CRF is used to identify Index lesions/tumors at baseline and to report the results of these Index lesions/tumors at all subsequent tumor assessment visits.
It illustrates the collection of the tumor identification information and the tumor results on a combined CRF. When SDTM submission datasets are created, the appropriate information must be mapped to TU and TR.
Tumor Identification/Result CRFs assume that measurable lesions that are followed and measured according to the criteria are classified as "Index lesions." All tumors that are followed but not measured according to the criteria are considered as "Non-Index lesions." These CRFs do not collect any tumor measurements for Non-Index lesions.
CDASH variable names for denormalized variables are examples, and are used to ensure unique CDASH variable names. Sponsors may use other conventions for creating denormalized CDASH variable names.
See Section 1.3, CDASH Metadata and Annotated CRFs, for explanation of annotations.
| Indicate whether or not Index lesions were identified. If Index lesions identified, list each tumor and provide the requested information. | Were tumors identified?
TUYN Not submitted |
<From NY codelist> |
|---|---|---|
| Sponsor specified |
TULNKID TULNKID and TRLNKID | _________________ |
| Specify the general (anatomical) location of the tumor. | What was the anatomical location of the tumor identified?
TULOC |
<From LOC codelist> |
| Specify the laterality of the tumor. | If applicable, what was the laterality of the anatomical location?
TULAT |
<From LAT codelist> |
| Specify the directionality of the tumor. | If applicable, what was the directionality of the anatomical location?
TUDIR |
<From DIR codelist> |
| Describe additional detail on the exact location of the tumor so that it can be distinguished from other tumors in the same anatomical location. |
TULOCDTL SUPPTU.QVAL where QNAM = "TULOCDTL" | _________________ |
| Indicate whether this is a tumor that has split from a previous tumor, or is a tumor that has merged with another tumor. | Select if changes to tumor were identified.
TUCHANGE If TUCHANGE = "Split" then TUORRES = "TARGET" where TUTESTCD = "TUSPLIT" If TUCHANGE = "Merged" then TUORRES = "TARGET" where TUTESTCD = "TUMERGE" |
<From TUTEST codelist> |
| Specify the method used to identify/evaluate the tumor. | What was the method of evaluation?
TUMETHOD TUMETHOD and TRMETHOD |
<From METHOD codelist> |
| Insert the date of the scan/image/examination (not the date on which it was read, or the visit date). |
TUDAT TUDTC and TRDTC | _ _ / _ _ _ / _ _ _ _ |
| Indicate who performed the assessment. | What was the role of the person performing the tumor evaluation?
TUEVAL TUEVAL and TREVAL |
<From EVAL codelist> |
| Identify the evaluator providing this evaluation. | What is the evaluator identifier?
TUEVALID TUEVALID and TREVALID |
<From MEDEVAL codelist> |
| Record the longest diameter of the tumor. |
LDIAM_TRORRES TRORRES where TRTESTCD = "LDIAM" | _____ |
|
LDIAM_TRORRESU TRORRESU where TRTESTCD = "LDIAM" Pre-populated | mm
<From UNIT codelist> | |
| Check if the longest diameter is too small to measure. | Check if the longest diameter was too small to measure.
TOOSMALL_LDIAM_TRORRES TRORRES = "TOO SMALL TO MEASURE" where TRTESTCD = "LDIAM" |
|
| Record the longest perpendicular diameter of the tumor. |
LPERP_TRORRES TRORRES where TRTESTCD = "LPERP" | _____ |
|
LPERP_TRORRESU TRORRESU where TRTESTCD = "LPERP" Pre-populated | mm
<From UNIT codelist> | |
| Check if the longest perpendicular diameter is too small to measure. | Check if the longest perpendicular diameter was too small to measure.
TOOSMALL_LPERP_TRORRES TRORRES = "TOO SMALL TO MEASURE" where TRTESTCD = "LPERP" |
|
| If appropriate, denote the tumor as inevaluable. | Check if the tumor was inevaluable.
TRINEVAL TRSTAT = "NOT DONE" where TRTESTCD = "TUMSTATE" |
|
| Indicate the reason why the tumor was inevaluable. | If the tumor was inevaluable, what was the reason?
TRREASND TRREASND where TRSTAT = "NOT DONE" and TRTESTCD = "TUMSTATE" | |
| For lymph node tumors only, denote whether the node is pathological or non-pathological. | What was the lymph node state?
LNSTATE_TRORRES TRORRES where TRTESTCD = "LNSTATE" |
|
CRF Metadata
Annotated CRF: Tumor Identification/Results - irRC - Non-Index Lesions
This CRF is only an example and is not meant to imply that any particular layout or collection plan is preferable over another.
This CRF is used to identify non-Index lesions/tumors at baseline and to report the results of these non-index lesions/tumors at all subsequent tumor assessment visits.
It illustrates the collection of the new tumor identification information and the new tumor results on a combined CRF. When SDTM submission datasets are created, the appropriate information must be mapped to TU and TR.
Tumor Identification/Result CRFs assume that new measurable lesions that are followed and measured according to the criteria are classified as "Index lesions." All new tumors that are followed but not measured according to the criteria are considered as "Non-Index lesions." These CRFs do not collect any tumor measurements for Non-Index lesions.
CDASH variable names for denormalized variables are examples, and are used to ensure unique CDASH variable names. Sponsors may use other conventions for creating denormalized CDASH variable names.
See Section 1.3, CDASH Metadata and Annotated CRFs, for explanation of annotations.
| Indicate whether or not non-index tumors were identified. If non-index tumors identified, list each tumor and provide the requested information. | Were tumors identified?
TUYN Not submitted |
<From NY codelist> |
|---|---|---|
| Sponsor specified |
TULNKID TULNKID and TRLNKID | _________________ |
| Specify the general (anatomical) location of the tumor. | What was the anatomical location of the tumor identified?
TULOC |
<From LOC codelist> |
| Specify the laterality of the tumor. | If applicable, what was the laterality of the anatomical location?
TULAT |
<From LAT codelist> |
| Specify the directionality of the tumor. | If applicable, what was the directionality of the anatomical location?
TUDIR |
<From DIR codelist> |
| Describe additional detail on the exact location of the tumor so that it can be distinguished from other tumors in the same anatomical location. |
TULOCDTL SUPPTU.QVAL where QNAM = "TULOCDTL" | _________________ |
| Used to specify non-measurable disease types that cannot be adequately described by anatomical location and other location qualifiers . | What was the tumor presentation type of the non-measurable disease?
TUPRTYP SUPPTU.QVAL |
<From DSPRTYP codelist> |
| Indicate whether this is a tumor that has split from a previous tumor, or is a tumor that has merged with another tumor. | Select if changes to tumor were identified.
TUCHANGE If TUCHANGE = "Split" then TUORRES = "NON-TARGET" where TUTESTCD = "TUSPLIT" If TUCHANGE = "Merged" then TUORRES = "NON-TARGET" where TUTESTCD = "TUMERGE" |
<From TUTEST codelist> |
| Specify the method used to identify/evaluate the tumor. | What was the method of evaluation?
TUMETHOD TUMETHOD and TRMETHOD |
<From METHOD codelist> |
| Insert the date of the scan/image/examination (not the date on which it was read, or the visit date). |
TUDAT TUDTC and TRDTC | _ _ / _ _ _ / _ _ _ _ |
| Indicate who performed the assessment. | What was the role of the person performing the tumor evaluation?
TUEVAL TUEVAL and TREVAL |
<From EVAL codelist> |
| Identify the evaluator providing this evaluation. | What is the evaluator identifier?
TUEVALID TUEVALID and TREVALID |
<From MEDEVAL codelist> |
| If appropriate, denote the tumor state. | What was the tumor state?
TUMSTATE_TRORRES TRORRES where TRTESTCD = "TUMSTATE" |
|
| If appropriate, denote the tumor as inevaluable. | Check if the tumor was inevaluable.
TRINEVAL TRSTAT = "NOT DONE" where TRTESTCD = "TUMSTATE" |
|
| Indicate the reason why the tumor was inevaluable. | If the tumor was inevaluable, what was the reason?
TRREASND TRREASND where TRSTAT = "NOT DONE" and TRTESTCD = "TUMSTATE" | |
| For lymph node tumors only, denote whether the node is pathological or non-pathological. | What was the lymph node state?
LNSTATE_TRORRES TRORRES where TRTESTCD = "LNSTATE" |
|
CRF Metadata
Annotated CRF: Tumor Identification/Results - irRC - New Lesions
This CRF is only an example and is not meant to imply that any particular layout or collection plan is preferable over another.
This CRF is used to identify new lesions/tumors identified during the study and to report the results of these new lesions/tumors.
It illustrates the collection of the new tumor identification information and the new tumor results on a combined CRF. When SDTM submission datasets are created, the appropriate information must be mapped to TU and TR.
Tumor Identification/Result CRFs assume that new measurable lesions that are followed and measured according to the criteria are classified as "Index lesions." All new tumors that are followed but not measured according to the criteria are considered "Non-Index lesions." These CRFs do not collect any tumor measurements for new Non-Index lesions.
CDASH variable names for denormalized variables are examples, and are used to ensure unique CDASH variable names. Sponsors may use other conventions for creating denormalized CDASH variable names.
See Section 1.3, CDASH Metadata and Annotated CRFs, for explanation of annotations.
| Indicate whether or not new tumors were identified. If new tumors identified, list each new tumor and provide the requested information. | Were new tumors identified?
TUYN Not submitted |
<From NY codelist> |
|---|---|---|
| Record or select which type of tumor is being evaluated as defined by the criteria. | What type of tumors are being identified as defined by the criteria being employed?
TUMIDENT_TUCORRES If TUMIDENT_TUCORRES = "New Index" then TUORRES = "NEW TARGET" where TUTESTCD = "TUMIDENT" If TUMIDENT_TUCORRES = "New Non-Index" then TUORRES = "NEW NON-TARGET" where TUTESTCD = "TUMIDENT" |
|
| Sponsor specified |
TULNKID TULNKID and TRLNKID | _________________ |
| Specify the general (anatomical) location of the tumor. | What was the anatomical location of the tumor identified?
TULOC |
<From LOC codelist> |
| Specify the laterality of the tumor. | If applicable, what was the laterality of the anatomical location?
TULAT |
<From LAT codelist> |
| Specify the directionality of the tumor. | If applicable, what was the directionality of the anatomical location?
TUDIR |
<From DIR codelist> |
| Describe additional detail on the exact location of the tumor so that it can be distinguished from other tumors in the same anatomical location. |
TULOCDTL SUPPTU.QVAL where QNAM = "TULOCDTL" | _________________ |
| Used to specify non-measurable disease types that cannot be adequately described by anatomical location and other location qualifiers | What was the tumor presentation type of the non-measurable disease?
TUPRTYP SUPPTU.QVAL where QNAM = "TUPRTYP" |
<From DSPRTYP codelist> |
| Indicate whether this is a tumor that has split from a previous tumor, or is a tumor that has merged with another tumor. | Select if changes to tumor were identified.
TUCHANGE If TUCHANGE = "Split" then TUORRES = TUMIDENT_TUORRES where TUTESTCD="TUSPLIT" If TUCHANGE = "Merged" then TUORRES = TUMIDENT_TUORRES where TUTESTCD = "TUMERGE" |
<From TUTEST codelist> |
| Specify the method used to identify/evaluate the tumor. | What was the method of evaluation?
TUMETHOD TUMETHOD and TRMETHOD |
<From METHOD codelist> |
| Insert the date of the scan/image/examination (not the date on which it was read, or the visit date). |
TUDAT TUDTC and TRDTC | _ _ / _ _ _ / _ _ _ _ |
| Indicate who performed the assessment. | What was the role of the person performing the tumor evaluation?
TUEVAL TUEVAL and TREVAL |
<From EVAL codelist> |
| Identify the evaluator providing this evaluation. | What is the evaluator identifier?
TUEVALID TUEVALID and TREVALID |
<From MEDEVAL codelist> |
| Record the longest diameter of the tumor. |
LDIAM_TRORRES TRORRES where TRTESTCD = "LDIAM" | _____ |
|
LDIAM_TRORRESU TRORRESU where TRTESTCD = "LDIAM" Pre-populated | mm
<From UNIT codelist> | |
| Check if the longest diameter is too small to measure. | Check if the longest diameter was too small to measure.
TOOSMALL_LDIAM_TRORRES TRORRES = "TOO SMALL TO MEASURE" where TRTESTCD = "LDIAM" |
|
| Record the longest perpendicular diameter of the tumor. |
LPERP_TRORRES TRORRES where TRTESTCD = "LPERP" | _____ |
|
LPERP_TRORRESU TRORRESU where TRTESTCD = "LPERP" Pre-populated | mm
<From UNIT codelist> | |
| Check if the longest perpendicular diameter is too small to measure. | Check if the longest perpendicular diameter was too small to measure.
TOOSMALL_LPERP_TRORRES TRORRES = "TOO SMALL TO MEASURE" where TRTESTCD = "LPERP" |
|
| If appropriate, denote the tumor state. | What was the tumor state?
TUMSTATE_TRORRES TRORRES where TRTESTCD = "TUMSTATE" |
|
| If appropriate, denote the tumor as inevaluable. | Check if the tumor was inevaluable.
TRINEVAL TRSTAT = "NOT DONE" where TRTESTCD = "TUMSTATE" |
|
| Indicate the reason why the tumor was inevaluable. | If the tumor was inevaluable, what was the reason?
TRREASND TRREASND where TRSTAT = "NOT DONE" and TRTESTCD = "TUMSTATE" | |
| For lymph node tumors only, denote whether the node is pathological or non-pathological. | What was the lymph node state?
LNSTATE_TRORRES TRORRES where TRTESTCD = "LNSTATE" |
|
CRF Metadata
Annotated CRF: Tumor Response - irRC
This CRF is only an example and is not meant to imply that any particular layout or collection plan is preferable over another.
This CRF is used to collect tumor responses during the study.
CDASH variable names for denormalized variables are examples, and are used to ensure unique CDASH variable names. Sponsors may use other conventions for creating denormalized CDASH variable names.
See Section 1.3, CDASH Metadata and Annotated CRFs, for explanation of annotations.
| Indicate whether or not response was collected. | Was the response assessment performed?
RSPERF If RSPERF = "No" then RSSTAT = "NOT DONE" where RSTESTCD = "OVRLRESP" |
<From NY codelist> |
|---|---|---|
| If the response was not collected, indicate why. | Why was the response assessment not performed?
RSREASND RSREASND where RSSTAT = "NOT DONE" and RSTESTCD = " OVRLRESP" |
|
| Indicate who performed the assessment. | Response Evaluator
RSEVAL |
<From EVAL codelist> |
| Identify the evaluator providing this evaluation. | What is the evaluator identifier?
RSEVALID |
<From MEDEVAL codelist> |
| Indicate the overall response assessment. | What was the Overall Response?
OVRLRESP_RSORRES RSORRES where RSTESTCD = "OVRLRESP" |
|
| Record the date of the procedure associated with the overall response. |
OVRLRESP_RSDAT RSDTC where RSTESTCD = "OVRLRESP" | _ _ / _ _ _ / _ _ _ _ |
| Indicate whether or not symptomatic deterioration was observed. | Did the patient experience Symptomatic Deterioration?
SYMPTDTR_RSORRES RSORRES where RSTESTCD = "SYMPTDTR" |
<From NY codelist> |
| Insert the date on which symptomatic deterioration was observed. |
SYMPTDTR_RSDAT RSDTC where RSTESTCD = "SYMPTDTR" | _ _ / _ _ _ / _ _ _ _ |
| Insert the name of the vendor performing the response assessments. |
RSNAM | _________________ |
| Indicate whether this evaluation is considered to be the accepted evaluation. | Was this record considered to be the accepted evaluation?
OVRLRESP_RSACPTFL RSACPTFL where RSTESTCD = " OVRLRESP" |
<From NY codelist> |
CRF Metadata
This example shows a representation of tumor assessments using irRC, which is based on bidimensional measurements following the WHO criteria[29]. The SDTM variable RSCAT is used to indicate which response criteria and version were used in a study.
Example
This is an example of tumors identified and tracked following iRC. In this example, the sponsor has chosen to represent the imaging procedure used for tumor evaluation in the Procedure (PR) domain. Imaging procedures were assigned --REFID. These --REFID were also represented in the TU, and TR datasets to link the each tumor assessment to the procedure used for assessment. The PR domain is not shown but examples have been provided in the TAUGs for Breast Cancer (Section 4.2) and Prostate Cancer (Section 4.2). This example primarily focuses on the modeling of key feature of the irRC criteria.
Subject 70001 had measurable and non-measurable tumors at baseline. In irRC (represented in RSCAT as "WOLCHOK SOLID TUMORS 2009"), tumor response is based on total measurable tumor burden and the development of new tumor(s) does not define progression. The tumor burden is defined by the sum of the products of the 2 largest perpendicular diameters (SUMPPD) of all index lesions (5 lesions per organ, up to 10 visceral lesions and 5 cutaneous index lesions) at baseline and new measurable tumors.
Although an Index lesion may be referred to as a Target Lesion and a Non-Index lesion may be referred to as a Non-Target lesion by some authors following RECIST language, the irRC criteria denote these tumors/lesions at baseline as Index or Non-Index lesions. Because the terms Index and Non-Index are synonymous with Target and Non-Target, respectively, the existing terminology values of "TARGET" and "NON-TARGET", with "INDEX" and "NON-INDEX" as synonyms, will be used in the published Controlled Terminology (CT). "NEW NON-TARGET" and "NEW TARGET" will also be added to the CT. In this study, the sponsor used the synonym terms "INDEX" and "NON-INDEX" on the CRF but used the terms in the CT for both TUORRES and TUSTRESC. The --GRPID variables were populated using Target, Non-Target, New Target, or New Non-Target. The --LNKID variables were populated using the notation that T=Target, NT=Non-Target, NEWT=New Target, and NEWNT=New Non-Target lesion.
At Assessment 2, the subject had 1 new Non-Target lesion. In irRC, Non-Target lesions are assessed as absent or present. At Assessment 3, 2 new Target lesions were identified.
| Rows 1-3: | Show that the subject had 2 measurable tumors (Index/Target lesions) in the right lung and a non-measurable tumor (Non-Index/Non-Target lesion) in the left lung at the screening assessment. Note that the standardized controlled terminology of "TARGET", and "NON-TARGET" are used to represent these lesions in TUORRES and TUSTRESC rather than the synonym terms used on the CRFs. |
|---|---|
| Row 4: | Shows the subject had a new non-measurable tumor identified (TULNKID="R-NEWNT01") in the liver at the Week 4 assessment. Note that the terminology "NEW NON-TARGET" is used in both TUORRES and TUSTRESC. |
| Rows 5-6: | Show the subject had 2 new measurable tumors identified (TULNKID="R-NEWT01 "and "R-NEWT02") in the left and right lung at the Week 8 assessment. Note that the terminology of "NEW TARGET" is used in TUORRES and TUSTRESC. |
tu.xpt
| Row | STUDYID | DOMAIN | USUBJID | TUSEQ | TUREFID | TULNKID | TUTESTCD | TUTEST | TUORRES | TUSTRESC | TUNAM | TULOC | TUMETHOD | TUEVAL | TUEVALID | VISITNUM | VISIT | EPOCH | TUDTC | TUDY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA123 | TU | 70001 | 1 | IMG-0002 | R-NT01 | TUMIDENT | Tumor Identification | NON-TARGET | NON-TARGET | ACME VENDOR | LUNG, LEFT UPPER LOBE | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | BASELINE | 2007-01-01 | 1 |
| 2 | CRCA123 | TU | 70001 | 2 | IMG-0003 | R-T01 | TUMIDENT | Tumor Identification | TARGET | TARGET | ACME VENDOR | LUNG, RIGHT LOWER LOBE | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | BASELINE | 2007-01-01 | 1 |
| 3 | CRCA123 | TU | 70001 | 3 | IMG-0004 | R-T02 | TUMIDENT | Tumor Identification | TARGET | TARGET | ACME VENDOR | LUNG, RIGHT MIDDLE LOBE | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | BASELINE | 2007-01-01 | 1 |
| 4 | CRCA123 | TU | 70001 | 4 | IMG-0008 | R-NEWNT01 | TUMIDENT | Tumor Identification | NEW NON-TARGET | NEW NON-TARGET | ACME VENDOR | LIVER, RIGHT LOBE | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 |
| 5 | CRCA123 | TU | 70001 | 5 | IMG-0012 | R-NEWT01 | TUMIDENT | Tumor Identification | NEW TARGET | NEW TARGET | ACME VENDOR | LUNG, LEFT LOWER LOBE | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 6 | CRCA123 | TU | 70001 | 6 | IMG-0013 | R-NEWT02 | TUMIDENT | Tumor Identification | NEW TARGET | NEW TARGET | ACME VENDOR | LUNG, RIGHT, SUPERIOR LOBE, ANTERIOR SEGMENT | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
This example TR dataset shows assessments needed to evaluate response using irRC guidelines. The assessments and the tumors that were identified and recorded in the TU domain are linked via tumor identifiers represented in TRLNKID. The TRGRPID variable is used to group the tumors. TRLNKGRP is used to organize the individual lesion assessments by disease assessment occasion. TRGRPID is not populated when summary results are presented (i.e., when TRTESTCD="SUMPPD', "PCBSPPD" and "PCNSPPD") as these summary results may be based on both Target and New Target lesions. The sponsor choose to create a separate, related PR record.
In this example, the sponsor has chosen to represent the imaging procedure information using the Procedure (PR) domain.
| Rows 1-6: | Show the results of the baseline assessments for each of the 2 identified Index lesions. The assessments are Longest Diameter, Longest Perpendicular, and Product of Perpendicular Diameters. |
|---|---|
| Row 7: | Shows the sum of the products of the perpendicular diameters. Because this assessment summarizes multiple lesions, TRLNKID is blank. |
| Row 8: | Shows that a non-measurable lesion (Non-Index) was present at baseline. |
| Rows 9-18: | Show the results for Assessment 2 (Week 4). Note that sum of product of the perpendicular diameters and the percent change in the sum of product of the perpendicular diameters from the baseline were reported. For the existing non-index and new non-index lesions, the assessment was "Tumor State" and the result was "PRESENT". |
| Rows 19-30: | Show measurement results for Target lesions at Assessment 3 (Week 8), including the target lesions identified at baseline and the 2 new target lesions identified at this assessment. |
| Rows 31-33: | Show the summary assessments for Assessment 3: the sum of product of the perpendicular diameters, the percent change of the sum of the products of the perpendicular diameters from baseline, and the percent change in the sum of the products of the perpendicular diameters from nadir. The nadir used to calculate percent change from nadir was observed at Assessment 2. Note that Index lesions and any New Index lesions are included in the sums of the areas. |
| Rows 34-35: | Show that both Non-Target lesions (non-index) are still present at Assessment 3. |
| Rows 36-52: | Show the results for Assessment 4. Assessment 4 was conducted to "confirm the irPD" observed in Assessment 3. |
tr.xpt
| Row | STUDYID | DOMAIN | USUBJID | TRSEQ | TRGRPID | TRREFID | TRLNKGRP | TRLNKID | TRTESTCD | TRTEST | TRORRES | TRORRESU | TRSTRESC | TRSTRESN | TRSTRESU | TRNAM | TRMETHOD | TREVAL | TREVALID | VISITNUM | VISIT | EPOCH | TRDTC | TRDY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA123 | TR | 70001 | 1 | TARGET | IMG-0003 | A1 | R-T01 | LDIAM | Longest Diameter | 10 | mm | 10 | 10 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 |
| 2 | CRCA123 | TR | 70001 | 2 | TARGET | IMG-0003 | A1 | R-T01 | LPERP | Longest Perpendicular | 5 | mm | 5 | 5 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 |
| 3 | CRCA123 | TR | 70001 | 3 | TARGET | A1 | R-T01 | PPD | Product of Perpendicular Diameters | 50 | mm2 | 50 | 50 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 | ||
| 4 | CRCA123 | TR | 70001 | 4 | TARGET | IMG-0004 | A1 | R-T02 | LDIAM | Longest Diameter | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 |
| 5 | CRCA123 | TR | 70001 | 5 | TARGET | IMG-0004 | A1 | R-T02 | LPERP | Longest Perpendicular | 10 | mm | 10 | 10 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 |
| 6 | CRCA123 | TR | 70001 | 6 | TARGET | A1 | R-T02 | PPD | Product of Perpendicular Diameters | 70 | mm2 | 70 | 70 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 | ||
| 7 | CRCA123 | TR | 70001 | 7 | A1 | SUMPPD | Sum of Products of Perpendicular Diam | 120 | mm2 | 120 | 120 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 | ||||
| 8 | CRCA123 | TR | 70001 | 8 | NON-TARGET | IMG-0002 | A1 | R-NT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 10 | SCREEN | SCREEN | 2007-01-01 | 1 | |||
| 9 | CRCA123 | TR | 70001 | 9 | TARGET | IMG-0005 | A2 | R-T01 | LDIAM | Longest Diameter | 10 | mm | 10 | 10 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 |
| 10 | CRCA123 | TR | 70001 | 10 | TARGET | IMG-0005 | A2 | R-T01 | LPERP | Longest Perpendicular | 5 | mm | 5 | 5 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 |
| 11 | CRCA123 | TR | 70001 | 11 | TARGET | A2 | R-T01 | PPD | Product of Perpendicular Diameters | 50 | mm2 | 50 | 50 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 | ||
| 12 | CRCA123 | TR | 70001 | 12 | TARGET | IMG-0006 | A2 | R-T02 | LDIAM | Longest Diameter | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 |
| 13 | CRCA123 | TR | 70001 | 13 | TARGET | IMG-0006 | A2 | R-T02 | LPERP | Longest Perpendicular | 9 | mm | 9 | 9 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 |
| 14 | CRCA123 | TR | 70001 | 14 | TARGET | A2 | R-T02 | PPD | Product of Perpendicular Diameters | 63 | mm2 | 63 | 63 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 | ||
| 15 | CRCA123 | TR | 70001 | 15 | A2 | SUMPPD | Sum of Products of Perpendicular Diam | 113 | mm2 | 113 | 113 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 | ||||
| 16 | CRCA123 | TR | 70001 | 16 | A2 | PCBSPPD | Percent Change Baseline in Sum of PPD | -5.8 | % | -5.8 | -5.8 | % | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 | ||||
| 17 | CRCA123 | TR | 70001 | 17 | NON -TARGET | IMG-0007 | A2 | R-NT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 | |||
| 18 | CRCA123 | TR | 70001 | 18 | NEW NON-TARGET | IMG-0008 | A2 | R-NEWNT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 20 | WEEK 4 | TREATMENT | 2007-01-28 | 28 | |||
| 19 | CRCA123 | TR | 70001 | 19 | TARGET | IMG-0009 | A3 | R-T01 | LDIAM | Longest Diameter | 12 | mm | 12 | 12 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 20 | CRCA123 | TR | 70001 | 20 | TARGET | IMG-0009 | A3 | R-T01 | LPERP | Longest Perpendicular | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 21 | CRCA123 | TR | 70001 | 21 | TARGET | A3 | R-T01 | PPD | Product of Perpendicular Diameters | 84 | mm2 | 84 | 84 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||
| 22 | CRCA123 | TR | 70001 | 22 | TARGET | IMG-0010 | A3 | R-T02 | LDIAM | Longest Diameter | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 23 | CRCA123 | TR | 70001 | 23 | TARGET | IMG-0010 | A3 | R-T02 | LPERP | Longest Perpendicular | 9 | mm | 9 | 9 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 24 | CRCA123 | TR | 70001 | 24 | TARGET | A3 | R-T02 | PPD | Product of Perpendicular Diameters | 63 | mm2 | 63 | 63 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||
| 25 | CRCA123 | TR | 70001 | 25 | NEW TARGET | IMG-0012 | A3 | R-NEWT01 | LDIAM | Longest Diameter | 10 | mm | 10 | 10 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 26 | CRCA123 | TR | 70001 | 26 | NEW TARGET | IMG-0012 | A3 | R-NEWT01 | LPERP | Longest Perpendicular | 5 | mm | 5 | 5 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 27 | CRCA123 | TR | 70001 | 27 | NEW TARGET | A3 | R-NEWT01 | PPD | Product of Perpendicular Diameters | 50 | mm2 | 50 | 50 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||
| 28 | CRCA123 | TR | 70001 | 28 | NEW TARGET | IMG-0013 | A3 | R-NEWT02 | LDIAM | Longest Diameter | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 29 | CRCA123 | TR | 70001 | 29 | NEW TARGET | IMG-0013 | A3 | R-NEWT02 | LPERP | Longest Perpendicular | 9 | mm | 9 | 9 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 |
| 30 | CRCA123 | TR | 70001 | 30 | NEW TARGET | A3 | R-NEWT02 | PPD | Product of Perpendicular Diameters | 63 | mm2 | 63 | 63 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||
| 31 | CRCA123 | TR | 70001 | 31 | A3 | SUMPPD | Sum of Products of Perpendicular Diam | 260 | mm2 | 260 | 260 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||||
| 32 | CRCA123 | TR | 70001 | 32 | A3 | PCBSPPD | Percent Change Baseline in Sum of PPD | 116.7 | % | 116.7 | 116.7 | % | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||||
| 33 | CRCA123 | TR | 70001 | 33 | A3 | PCNSPPD | Percent Change Nadir in Sum of PPD | 130.1 | % | 130.1 | 130.1 | % | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | ||||
| 34 | CRCA123 | TR | 70001 | 34 | NON-TARGET | IMG-0015 | A3 | R-NT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | |||
| 35 | CRCA123 | TR | 70001 | 35 | NEW NON-TARGET | IMG-0016 | A3 | R-NEWNT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 30 | WEEK 8 | TREATMENT | 2007-02-25 | 56 | |||
| 36 | CRCA123 | TR | 70001 | 36 | TARGET | IMG-0017 | A4 | R-T01 | LDIAM | Longest Diameter | 12 | mm | 12 | 12 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 37 | CRCA123 | TR | 70001 | 37 | TARGET | IMG-0017 | A4 | R-T01 | LPERP | Longest Perpendicular | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 38 | CRCA123 | TR | 70001 | 38 | TARGET | A4 | R-T01 | PPD | Product of Perpendicular Diameters | 84 | mm2 | 84 | 84 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||
| 39 | CRCA123 | TR | 70001 | 39 | TARGET | IMG-0018 | A4 | R-T02 | LDIAM | Longest Diameter | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 40 | CRCA123 | TR | 70001 | 40 | TARGET | IMG-0018 | A4 | R-T02 | LPERP | Longest Perpendicular | 9 | mm | 9 | 9 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 41 | CRCA123 | TR | 70001 | 41 | TARGET | A4 | R-T02 | PPD | Product of Perpendicular Diameters | 63 | mm2 | 63 | 63 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||
| 42 | CRCA123 | TR | 70001 | 42 | NEW TARGET | IMG-0019 | A4 | R-NEWT01 | LDIAM | Longest Diameter | 10 | mm | 10 | 10 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 43 | CRCA123 | TR | 70001 | 43 | NEW TARGET | IMG-0019 | A4 | R-NEWT01 | LPERP | Longest Perpendicular | 5 | mm | 5 | 5 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 44 | CRCA123 | TR | 70001 | 44 | NEW TARGET | A4 | R-NEWT01 | PPD | Product of Perpendicular Diameters | 50 | mm2 | 50 | 50 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||
| 45 | CRCA123 | TR | 70001 | 45 | NEW TARGET | IMG-0020 | A4 | R-NEWT02 | LDIAM | Longest Diameter | 7 | mm | 7 | 7 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 46 | CRCA123 | TR | 70001 | 46 | NEW TARGET | IMG-0020 | A4 | R-NEWT02 | LPERP | Longest Perpendicular | 9 | mm | 9 | 9 | mm | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
| 47 | CRCA123 | TR | 70001 | 47 | NEW TARGET | A4 | R-NEWT02 | PPD | Product of Perpendicular Diameters | 63 | mm2 | 63 | 63 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||
| 48 | CRCA123 | TR | 70001 | 48 | A4 | SUMPPD | Sum of Products of Perpendicular Diam | 260 | mm2 | 260 | 260 | mm2 | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||||
| 49 | CRCA123 | TR | 70001 | 49 | A4 | PCBSPPD | Percent Change Baseline in Sum of PPD | 116.7 | % | 116.7 | 116.7 | % | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||||
| 50 | CRCA123 | TR | 70001 | 50 | A4 | PCNSPPD | Percent Change Nadir in Sum of PPD | 130.1 | % | 130.1 | 130.1 | % | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | ||||
| 51 | CRCA123 | TR | 70001 | 51 | NON -TARGET | IMG-0021 | A4 | R-NT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 | |||
| 52 | CRCA123 | TR | 70001 | 52 | NEW NON-TARGET | IMG-0022 | A4 | R-NEWNT01 | TUMSTATE | Tumor State | PRESENT | PRESENT | ACME VENDOR | CT SCAN | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | 40 | WEEK 12 | TREATMENT | 2007-03-25 | 84 |
This example shows the tumor responses associated with the assessments provided in the TR domain using irRC. The (ONCRSR) codelist is used to report tumor response. The values of irCR, irPR, irSD, and irPD were used to collect the tumor response. These values were also used by the sponsor in the SDTM-based dataset as this codelist is extensible. It is recommended that the overall response of confirmed irPD, as well as the associated dates of these events, be derived in ADaM based on the individual assessor's responses collected at each assessment. For example, in the collected data and in the SDTM-based dataset, Overall Responses are reported as irPD, without indicating whether this is an unconfirmed or confirmed irPD. This approach was considered the best option to avoid discrepancies between assessor's responses (collected data) and derived ADaM data. Using this approach, the sponsor ensures that the protocol rules for defining a confirmed irPD and the date of the confirmed irPD are applied. The sponsor usually has very detailed analysis derivations for these key endpoints and may conduct several sensitivity analyses to handle special cases. These rules can be about the frequency of assessments as well as missing assessments and missing evaluation on tumors being followed.
| Row 1: | Shows the overall response according to irRC guidelines at Assessment 2. The observed percent change from baseline in sum of areas was a decrease of 5.8%, which was neither a 50% decrease nor a 25% increase, so the overall response was stable disease. This is represented in RSORRES and RSSTRESC as "irSD". Under the irRC guidelines, the presence of the new non-measurable tumor is not considered progressive disease. |
|---|---|
| Row 2: | Shows the irRC overall response using irRC guidelines at Assessment 3. Because the observed percent change from nadir in sum of areas was an increase of 130.1%, the overall response was progressive disease which has not yet been confirmed. This is represented in RSORRES and RSSTRESC as "irPD". |
| Row 3: | Shows the irRC overall response using irRC guidelines at Assessment 4. The observed percent change from nadir in sum of areas was an increase of 130.1%. Although Assessment 4 was conducted to confirm the progressive disease observed at Assessment 3 and the same overall response was observed at Assessment 4, the result of the overall assessment is still recorded as "irPD" because an overall response of confirmed irPD will be derived in ADaM according to the rules defined in the protocol for this study. This is represented in RSORRES and RSSTRESC as "irPD". |
rs.xpt
| Row | STUDYID | DOMAIN | USUBJID | RSSEQ | RSLNKGRP | RSTESTCD | RSTEST | RSCAT | RSORRES | RSSTRESC | RSNAM | RSEVAL | RSEVALID | EPOCH | VISITNUM | VISIT | RSDTC | RSDY |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | CRCA123 | RS | 70001 | 1 | A2 | OVRLRESP | Overall Response | WOLCHOK SOLID TUMORS 2009 | irSD | irSD | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | TREATMENT | 20 | WEEK 4 | 2007-01-28 | 28 |
| 2 | CRCA123 | RS | 70001 | 2 | A3 | OVRLRESP | Overall Response | WOLCHOK SOLID TUMORS 2009 | irPD | irPD | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | TREATMENT | 30 | WEEK 8 | 2007-02-26 | 56 |
| 3 | CRCA123 | RS | 70001 | 3 | A4 | OVRLRESP | Overall Response | WOLCHOK SOLID TUMORS 2009 | irPD | irPD | ACME VENDOR | INDEPENDENT ASSESSOR | RADIOLOGIST 1 | TREATMENT | 40 | WEEK 12 | 2007-03-26 | 84 |
An example of the dataset relationships (RELREC) is not shown as it has been described in the SDTMIG.
5 Questionnaires, Ratings, and Scales
Colorectal cancer studies may use measures which assess symptoms, a particular symptom such as functional status (i.e., ability to perform functions of daily living), or health-related quality of life.
Questionnaires, Ratings, and Scales (QRS) are maintained as standalone guides on the CDISC website: https://www.cdisc.org/foundational/qrs. The table below lists assessments that are being pursued as potential supplements as part of the development work for this TAUG. Supplements may or may not be finalized at the time of publication, and depend on copyright approval where applicable. CDISC cannot produce supplements for copyrighted measures without the express permission of the copyright holder.
Sponsors should refer to QRS guides on the CDISC website for measures of interest not included below, as they may have been developed for another therapeutic area; new measures are implemented on an ongoing basis by the CDISC QRS Terminology and Standards Development sub-teams. See CDISC COP 001 (https://www.cdisc.org/about/bylaws) for details on implementing or requesting development of standards for SDTM-based submissions.
Identified QRS Measures of Interest to Colorectal Cancer
| Full Name and Abbreviation | Copyright Permission Status | Supplement Status |
|---|---|---|
| Eastern Cooperative Oncology Group-Performance Status (ECOG) | Granted | Done |
| Karnofsky Performance Status (KPS SCALE) | Granted | Done |
| EORTC QLQ-CR29 | Granted | In progress |
| EORTC QLQ-C30 | Granted | In progress |
| EORTC QLQ-LMC21 | Requested | |
| Functional Assessment of Cancer Therapy - Colorectal (Version 4) (FACT-C) | Granted | In progress |
| European Quality of Life Five Dimension Five Level Scale (EQ-5D-5L) | Granted | Done |
| European Quality of Life Five Dimension Three Level Scale (EQ-5D-3L) | Granted | Done |
6 Routine Data
Some routinely collected data are not specific to a therapeutic area. These include data collected to assess the subject's history, the safety of the study treatment, and/or factors in assessing the subject's condition or reaction to a treatment.
For many cancer treatments, one of the aims of early development is the characterization of side effects, including the identification of dose-limiting toxicities. This aspect of colorectal cancer study data is beyond the scope of this version of the TAUG.
In later development, when considering adverse events that have been found to be associated with a drug, data collection will depend on the particular side effects. As this varies considerably among cancer treatments, it is not covered here. Advice for analysis of adverse events within ADaM can be found in the ADaM Structure for Occurrence Data (OCCDS) available at https://www.cdisc.org/standards/foundational/adam.
Pre-specified adverse events that may be of interest but often depend on the treatment given. Advice for collecting data on pre-specified adverse events may be found in the CDASH and SDTM foundational standards.
Medications may be given for pain management. Time to first use of opiates may be used as an endpoint in a study. CDISC forming a team that will focus on the SDTM modeling of combination products/regimens. This topic is applicable to many therapeutic areas. CDISC anticipates that this modeling will be published in a stand-alone publication and would provide guidelines for the representation of combination products or combination regimens.
7 Analysis Data
This section presents some of the concepts and common analysis endpoints relevant to the analysis of colorectal cancer studies. Colorectal cancer is considered a solid tumor. Clinical trial data would be analyzed like other clinical studies in solid tumors (e.g., breast cancer, prostate cancer). Therefore, most of the methodologies described in those TAUGs would be the same for colorectal cancer.
7.1 Analysis Endpoints
Colorectal cancer efficacy analysis mainly falls into two groups: response analysis and time-to-event analysis. Typical time-to-event endpoints are:
- Progression-Free Survival (PFS)
- Overall Survival (OS)
- Disease-Free Survival (DFS)
Other time-to-event endpoints may be considered (e.g., cancer-specific survival) depending on the protocol planned analyses.
The methodologies for creating datasets to support these analyses have been covered in previous TAUGs (Breast Cancer and Prostate Cancer).
Best Overall Response (BOR) analysis for colorectal cancer studies is based on RECIST 1.1, irRC, irRECIST, or iRECIST. BOR is defined as the best response recorded from the start of the trial—normally the date of randomization—until death, until disease progression occurs either by RECIST or irRC, or until the patient discontinues treatment. BOR analysis based on RECIST criteria is described in the BrCa and PrCa TAUGs.
The immune related response criteria (irRC)[28] has been developed to assess response and progression where patients are treated with an immune-oncology drug. RECIST criteria, which are typically used to assess solid tumor cancers, are not adequate because they do not account for the time gap in many patients between initial treatment and the apparent action of the immune system to reduce the tumor burden.
In metastatic colon cancer clinical trials irRC is emerging as a method to evaluate efficacy endpoints.
Some of the differences between RECIST and irRC are:
- RECIST looks at "Target" and "Non-target" lesions whereas irRC looks at "Index" and "Non-index" lesions.
- In irRC, new lesions are considered a change in tumor burden and not necessarily progressive disease, whereas in RECIST a new lesion is considered to be a sign of disease progression.
- Lesions in RECIST are measured unidimensionally based on the longest diameter whereas lesions in irRC are measured bidimensionally.
- Algorithms for determining responses (including progression) differ between RECIST and irRC.
Documenting new lesion measurements is especially important in irRC. Whereas in RECIST new lesions always constitute progression, in irRC new index lesion measurements are included with the other index lesions measurements to determine tumor burden, and do not always constitute progression. This is due to the specific reaction (pseudo progression) to the immunologic drugs that may occur initially and subside in the subsequent assessments.
The response values for RECIST versus irRC are similar and are typically analyzed in the same way. When performing a best overall response analysis, the ranking for best response is considered to be complete response (CR/irCR), partial response (PR/irPR), stable disease (SD/irSD; Non-CR/Non-PD for subjects without target or index lesions), and progressive disease (PD/irPD), in that order. BOR can be calculated either for the entire time a subject was in the trial or for a specific period of time, such as for the combination therapy period and the maintenance therapy period.
Unlike the survival analysis, BOR analysis is based on tumor assessments performed until progressive disease is determined. Considerations for missing assessments are not generally taken into account. The BOR is analyzed based on the number of subjects achieving each of the RECIST or/and irRC categories during the defined clinical trial period.
7.2 Additional Concepts Related to Analysis
Almost all oncology studies will include the stage of cancer for each subject at the time the subject was included in the study. The most commonly used colorectal cancer staging system is known as the TNM system, established by the American Joint Committee on Cancer (AJCC). This system looks at T (tumor expansion), N (extent of cancer spread to lymph nodes), and M (metastases or spread of cancer to other organs). In addition to TNM information, colorectal cancer trials should consider collecting anatomical location details of the primary tumor and the distance from anal verge to the primary tumor. Anal sphincter preservation status at the time of the original surgery may also be of interest.
The performance of 2 types of surgical intervention, primary tumor resection and resection of liver metastases, may be important for efficacy analysis of colorectal cancer; information related to these procedures is often collected in colorectal cancer studies. If the primary tumor is rectal, information on whether total mesorectal excision (TME) was performed can also be important.
The following subject-level values should be considered to be captured for analysis:
- A flag to indicate the finding of No Existing Disease (NED) during the surgery
- The date of the surgical NED (there could be more than one surgery performed)
Performance status at baseline should be captured; the Eastern Cooperative Oncology Group-Performance Status (ECOG), Karnofsky Performance Status, and WHO Performance Status measures are all different methodologies for recording performance status.[33,34]
Some colorectal cancer characteristics that are considered risk factors include obstruction or perforation of the bowel, lymphovascular invasion, poorly differentiated histology, and number of resected lymph nodes. Other risk factors may also be assessed. The following are specific tests that assess some of these risk factors; all of these tests are used in the adjuvant setting only:
- Oncotype DX, which gives a score that measures risk of recurrence of colorectal cancer
- ColoPrint, which predicts Stage II recurrence of colorectal cancer
- CDX2 expression, which is a marker for gastrointestinal differentiation in colorectal cancer
Prior therapy information can also serve as a stratification indicator. Some of the interventions that might be captured are surgical interventions, non-surgical liver directed intervention, systemic therapy, and radiation. The PrCa TAUG has examples of variables that indicate types of intervention and important dates associated with these therapies.
7.2.1 Disease Characteristics
Disease characteristics used for prognostic stratification or covariates may be included in the subject-level analysis dataset (ADSL). Examples of disease characteristics that might be included in ADSL include mutations, microsatellite instability, and gene expression as determined by immunohistochemistry (IHC).
Mutation
Collecting the mutation type and subtype associated with a tumor specimen can help predict responses to EGFR-targeted immunotherapies. Method of mutation detection would also be important.
Microsatellite Instability (MSI)
Cancers with MSI account for approximately 15% of all colorectal cancers.[14] However, in some geographical regions (e.g., Japan) this may be lower. A tumor is classified as MSI-High if 2 or more of the 5 microsatellite sequences have been mutated and MSI-Low if only 1 has mutated. Detection of MSI is important to diagnose Lynch syndrome as well as to determine an effective chemotherapy regimen. MSI tumors do not respond well to 5-fluorouracil (5-FU), which is the most common treatment for colorectal cancer. Testing for MSI mutations might also be performed for prognostic stratification. This might be captured in ADSL.
Immunohistochemistry (IHC)
Immunohistochemistry (IHC) is a diagnostic technique used to identify discrete tissue components in a tissue specimen. Using specific tumor markers, IHC can be used to diagnose a cancer as benign or malignant, determine the stage and grade of a tumor, and identify the cell type and origin of a metastasis to find the site of the primary tumor. It also allows detection of expression of certain protein markers in a tumor which can be a prognostic factor in treatment and expected outcome to the disease. Over-expression of a protein may or may not indicate a gene mutation. The protein parameters may be target protein detection, intensity score, and percent cell staining. Intensity score and percent staining can be used to derive the outcome of the test as negative, positive, or other.
7.2.2 Carcinoembryonic Antigen (CEA)
Carcinoembryonic Antigen (CEA) is an important indicator in colorectal cancer trials. CEA levels at the baseline and throughout a particular study can indicate how widespread the cancer is as well indicating the success or lack thereof of the treatment. It can also be used to identify recurrences after surgical resection.
Appendices
Appendix A: Colorectal Cancer Team
| Name | Institution/Organization |
|---|---|
| John Owen, Team Lead | CDISC |
| Stephen Abel | Abbvie |
| Peter Ansell | Abbvie |
| Glenn Barnes | Takeda |
| Dana Booth | CDISC |
| Julie Chason | CDISC |
| Anthony Chow | CDISC |
| Christine Connolly | EMD Serono |
| Tara Erb | Lilly |
| John Ezzell | CDISC |
| Nate Freimark | The Griesser Group |
| Iska Gans | Roche |
| Marie-Laurence Harle-Yge | Roche |
| Joyce Hernandez | Independent |
| James Kramer | JnJ |
| Elizabeth Langevin | Takeda |
| Barbara Leutgeb | Roche |
| Kathleen Mellars | CDISC |
| Monica Motwani | Abbvie |
| Barbara Mueller | Roche |
| Erin Muhlbradt | NCI EVS |
| Catherine Mulvaney | Imaging Endpoints |
| Federico Nasroulah | Lilly |
| Anh Nguyen | Abbvie |
| Melanie Paules | GSK |
| Liao Qiming | Abbvie |
| Qin Qin | Abbvie |
| Dianne Reeves | NIH (National Institutes of Health) |
| Ellen Schatz | Lilly |
| Stefan Sherer | Novartis |
| Barbara A. Sowinski | Pfizer |
| Alana St. Clair | CDISC |
| Anita Umesh | Illumina |
| Darcy Wold | CDISC |
| Diane Wold | CDISC |
Appendix B: Non-Standard Variables (NSVs)
The following table lists the non-standard variables used in this document, and gives their parent domain and variable-level metadata. Note that names of the NSVs are listed using the -- notation to represent the 2 letter domain name abbreviations. These NSVs may be used in other domains, if appropriate.
Appendix C: Glossary and Abbreviations
| aCRF | annotated case report form |
| ADaM | Analysis Data Model |
| ADaMIG | ADaM Implementation Guide |
| BRIDG | Biomedical Research Integrated Domain Group |
| Biomedical concept | A high-level building block of clinical research and/or healthcare information that encapsulates lower-level implementation details like variables and terminologies. |
| CDASH | Clinical Data Acquisition Standards Harmonization Project |
| CDISC | Clinical Data Interchange Standards Consortium |
| CFAST | Coalition for Accelerating Standards and Therapies |
| Collected | "Collected" refers to information that is recorded and/or transmitted to the sponsor. This includes data entered by the site on CRFs/eCRFs as well as vendor data such as core lab data. This term is a synonym for "captured." |
| CT | Computed Tomography |
| Controlled Terminology | A finite set of values that represent the only allowed values for a data item. These values may be codes, text, or numeric. A code list is one type of controlled terminology. |
| CRF | Case report form (sometimes called a case record form). A printed, optical, or electronic document designed to record all required information to be reported to the sponsor for each trial subject. |
| Domain | A collection of observations with a topic-specific commonality about a subject. |
| ECOG | Eastern Cooperative Oncology Group |
| EGFR | Epidermal growth factor receptor |
| FISH | Fluorescence in situ hybridization |
| Foundational Standards | Used to refer to the suite of CDISC standards that describe the clinical study protocol (Protocol), design (Study Design), data collection (CDASH), laboratory work (Lab), analysis (ADaM), and data tabulation (SDTM and SEND). See https://www.cdisc.org for more information on each of these clinical data standards. |
| IHC | Immunohistochemistry |
| irRC | Immune-related Response Criteria based on WHO (WOLCHOK SOLID TUMORS 2009) |
| MRI | Magnetic resonance imaging |
| NCI | National Cancer Institute |
| NSV | Non-Standard Variable |
| OCCDS | ADaM Structure for Occurrence Data |
| Patient | A recipient of medical treatment. |
| PET | Positron Emission Tomography |
| PRO | Patient-reported outcome |
| RECIST | Response Evaluation Criteria in Solid Tumors |
| RT-qPCR | Quantitative reverse transcription polymerase chain reaction |
| SDS | Submission Data Standards. Also the name of the team that maintains the SDTM and SDTMIG. |
| SDTM | Study Data Tabulation Model |
| SDTMIG | SDTM Implementation Guide (for Human Clinical Trials) |
| SDTMIG-PGx | Study Data Tabulation Model Implementation Guide for Pharmacogenomics and Pharmacogenetics |
| SPD | Sum of the products of the two largest perpendicular diameters |
| Subject | A participant in a study. |
| WHO | World Health Organization |
Appendix D: References
Works Cited
- GLOBOCAN 2012: Colorectal cancer estimated incidence, mortality and prevalence worldwide in 2012. World Health Organization, International Agency for Research on Cancer. ( http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx?cancer=colorectal). Accessed October 10, 2017.
- Bowel cancer: Surger for advanced cancer. Cancer Research UK. ( https://www.cancerresearchuk.org/about-cancer/bowel-cancer/advanced/treatment/surgery ). Accessed October 27, 2016.
- Nivatvongs S, Rojanasakul A, Reiman HM, et al. The risk of lymph node metastasis in colorectal polyps with invasive adenocarcinoma. Dis Colon Rectum. 1991;34(4):323-328. doi:10.1007/BF02050592.
- Katoh H, Yamashita K, Wang G, Sato T, Nakamura T, Watanabe M. Prognostic significance of preoperative bowel obstruction in stage III colorectal cancer. Ann Surg Oncol. 2011;18(9):2432-2441. doi:10.1245/s10434-011-1625-3.
- Ogawa M, Watanabe M, Eto K, et al. Clinicopathological features of perforated colorectal cancer. Anticancer Res. 2009;29(5):1681-1684.
- Duffy MJ. Carcinoembryonic antigen as a marker for colorectal cancer: is it clinically useful? Clin Chem. 2001;47(4):624-630.
- Park IJ, Choi GS, Jun SH. Prognostic value of serum tumor antigen CA19-9 after curative resection of colorectal cancer. Anticancer Res. 2009;29(10):4303-4308.
- Koukourakis MI, Giatromanolaki A, Sivridis E, et al. Prognostic and predictive role of lactate dehydrogenase 5 expression in colorectal cancer patients treated with PTK787/ZK 222584 (vatalanib) antiangiogenic therapy. Clin Cancer Res. 2011;17(14):4892-4900. doi:10.1158/1078-0432.CCR-10-2918.
- Marmorino F, Salvatore L, Barbara C, et al. Serum LDH predicts benefit from bevacizumab beyond progression in metastatic colorectal cancer. Br J Cancer. 2017;116(3):318-323.
- Dalerba P, Sahoo D, Paik S, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med. 2016;374(3):211-222. doi:10.1056/NEJMoa1506597.
- Genetics of colorectal cancer (PDQ®) - health professional version. Executive summary. National Cancer Institute Web site. (https://www.cancer.gov/types/colorectal/hp/colorectal-genetics-pdq). Updated June 14, 2018. Accessed September 14, 2018.
- Schmoll HJ. Targeting HER2: precision oncology for colorectal cancer. Lancet Oncol. 2016;17(6):685-686. doi:10.1016/S1470-2045(16)30039-0.
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17(6):738-746. doi:10.1016/S1470-2045(16)00150-9.
- Horvat M, Stabuc B. Microsatellite instability in colorectal cancer. Radiol Oncol. 2011;45(2):75-81. doi:10.2478/v10019-011-0005-8.
- Nazemalhosseini Mojarad E, Kashfi SM, Mirtalebi H, et al. Low level of microsatellite instability correlates with poor clinical prognosis in stage II colorectal cancer patients. J Oncol. 2016;2016:2196703. doi:10.1155/2016/2196703.
- Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 2010;51:71-119.
- De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753-762. doi:10.1016/S1470-2045(10)70130-3.
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26(35):5705-5712. doi:10.1200/JCO.2008.18.0786.
- Semrad TJ, Kim EJ. Molecular testing to optimize therapeutic decision making in advanced colorectal cancer. J Gastrointest Oncol. 2016;7(Suppl 1):S11-20. doi:10.3978/j.issn.2078-6891.2015.094.
- Fodde R. The APC gene in colorectal cancer. Eur J Cancer. 2002;38(7):867-871.
- Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11(1):35-41. doi:10.1097/GIM.0b013e31818fa2ff.
- Libutti SK, Willett CG, Saltz LB. Cancer of the rectum. In: DeVita VT, Lawrence TS, Rosenberg SA, eds. Cancer: Principles and Practice of Oncology. 9th ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2011:1127-1141.
- Den dunnen JT, Dalgleish R, Maglott DR, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37(6):564-569. doi:10.1002/humu.22981.
- Adenocarcinoma (code C2852). National Cancer Institute Thesaurus. (https://ncit.nci.nih.gov/). Accessed September 14, 2018.
- Colorectal cancer survival linked to primary tumor location. National Cancer Institute. ( https://www.cancer.gov/news-events/cancer-currents-blog/2016/colorectal-survival-location ). Published May 27, 2016. Accessed September 14, 2018.
- Xiao Y, Wu B2, Qiu H, et al. Effects of tumor distance from anal verge and types of operations on survival outcomes for low rectal cancer after neoadjuvant chemoradiotherapy [in Chinese]. Zhonghua Yi Xue Za Zhi. 2014;94(22):1705-1709.
- Restivo A, Zorcolo L, Cocco IM, et al. Elevated CEA levels and low distance of the tumor from the anal verge are predictors of incomplete response to chemoradiation in patients with rectal cancer. Ann Surg Oncol. 2013;20(3):864-871. doi:10.1245/s10434-012-2669-8.
- Wolchok JD, Hoos A, O'Day S, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clin Cancer Res. 2009;15(23):7412-7420. doi:10.1158/1078-0432.CCR-09-1624.
- Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–214.
- Nishino M, Giobbie-hurder A, Gargano M, Suda M, Ramaiya NH, Hodi FS. Developing a common language for tumor response to immunotherapy: immune-related response criteria using unidimensional measurements. Clin Cancer Res. 2013;19(14):3936-3943. doi:10.1158/1078-0432.CCR-13-0895.
- Bohnsack O, Hoos A, Ludajic K. Adaptation of the immune related response criteria: irRECIST. Ann Oncol. 2014;25(suppl 4):iv361-iv372. doi:10.1093/annonc/mdu342.23.
- Seymour L, Bogaerts J, Perrone A, et al. iRECIST: guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017;18(3):e143-e152. doi:10.1016/S1470-2045(17)30074-8.
- Oken M, Creech R, Tormey D, et al. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649-655.
- Karnofsky DA, Burchenal JH. The Clinical Evaluation of Chemotherapeutic Agents in Cancer. In: MacLeod CM, ed. Evaluation of Chemotherapeutic Agents. New York: Columbia Univ Press; 1949.
Further Reading
-
Bartley AN, Hamilton SR, Alsabeh R, et al. Template for reporting results of biomarker testing of specimens from patients with carcinoma of the colon and rectum. Arch Pathol Lab Med. 2014;138(2):166-170. doi:10.5858/arpa.2013-0231-CP.
-
Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45(2):228-247. doi:10.1016/j.ejca.2008.10.026.
- Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29(35):4611-4619. doi:10.1200/JCO.2010.32.8732
- Dalerba P, Sahoo D, Paik S, et al. CDX2 as a prognostic biomarker in stage II and stage III colon cancer. N Engl J Med. 2016;374(3):211-222. doi:10.1056/NEJMoa1506597
-
IUPAC-IUB Joint Commission on Biochemical Nomenclature. Nomenclature and symbolism for amino acids and peptides. Recommendations 1983. Pure Appl. Chem. 1984;56(5):595-624. doi:10.1351/pac198456050595.
-
Marmorino F, Salvatore L, Barbara C, et al. Serum LDH predicts benefit from bevacizumab beyond progression in metastatic colorectal cancer. Br J Cancer. 2017;116(3):318-323.
- Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29(1):17-24. doi:10.1200/JCO.2010.30.1077
-
Umar A, Boland CR, Terdiman JP, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261-268.
- US Department of Health and Human Services, Food and Drug Administration, CDER. Guidance for Industry: Clinical Trial Endpoints for the Approval of Cancer Drugs and Biologics. Silver Spring, MD: U.S. Food and Drug Administration; 2007. ( http://www.fda.gov/downloads/drugs/guidancecomplianceregulatoryinformation/guidances/ucm071590.pdf). Accessed October 27, 2016.
-
Wain HM, Bruford EA, Lovering RC, Lush MJ, Wright MW, Povey S. Guidelines for human gene nomenclature. Genomics. 2002;79(4):464-470. doi:10.1006/geno.2002.6748.
-
World Health Organization. WHO handbook for reporting results of cancer treatment. Geneva, Switzerland: World Health Organization; 1979. WHO offset publication no. 48. ( http://www.who.int/iris/handle/10665/37200 ). Accessed September 5, 2018.
Appendix E: Representations and Warranties, Limitations of Liability, and Disclaimers
CDISC Patent Disclaimers
It is possible that implementation of and compliance with this standard may require use of subject matter covered by patent rights. By publication of this standard, no position is taken with respect to the existence or validity of any claim or of any patent rights in connection therewith. CDISC, including the CDISC Board of Directors, shall not be responsible for identifying patent claims for which a license may be required in order to implement this standard or for conducting inquiries into the legal validity or scope of those patents or patent claims that are brought to its attention.
Representations and Warranties
"CDISC grants open public use of this User Guide (or Final Standards) under CDISC's copyright."
Each Participant in the development of this standard shall be deemed to represent, warrant, and covenant, at the time of a Contribution by such Participant (or by its Representative), that to the best of its knowledge and ability: (a) it holds or has the right to grant all relevant licenses to any of its Contributions in all jurisdictions or territories in which it holds relevant intellectual property rights; (b) there are no limits to the Participant's ability to make the grants, acknowledgments, and agreements herein; and (c) the Contribution does not subject any Contribution, Draft Standard, Final Standard, or implementations thereof, in whole or in part, to licensing obligations with additional restrictions or requirements inconsistent with those set forth in this Policy, or that would require any such Contribution, Final Standard, or implementation, in whole or in part, to be either: (i) disclosed or distributed in source code form; (ii) licensed for the purpose of making derivative works (other than as set forth in Section 4.2 of the CDISC Intellectual Property Policy ("the Policy")); or (iii) distributed at no charge, except as set forth in Sections 3, 5.1, and 4.2 of the Policy. If a Participant has knowledge that a Contribution made by any Participant or any other party may subject any Contribution, Draft Standard, Final Standard, or implementation, in whole or in part, to one or more of the licensing obligations listed in Section 9.3, such Participant shall give prompt notice of the same to the CDISC President who shall promptly notify all Participants.
No Other Warranties/Disclaimers. ALL PARTICIPANTS ACKNOWLEDGE THAT, EXCEPT AS PROVIDED UNDER SECTION 9.3 OF THE CDISC INTELLECTUAL PROPERTY POLICY, ALL DRAFT STANDARDS AND FINAL STANDARDS, AND ALL CONTRIBUTIONS TO FINAL STANDARDS AND DRAFT STANDARDS, ARE PROVIDED "AS IS" WITH NO WARRANTIES WHATSOEVER, WHETHER EXPRESS, IMPLIED, STATUTORY, OR OTHERWISE, AND THE PARTICIPANTS, REPRESENTATIVES, THE CDISC PRESIDENT, THE CDISC BOARD OF DIRECTORS, AND CDISC EXPRESSLY DISCLAIM ANY WARRANTY OF MERCHANTABILITY, NONINFRINGEMENT, FITNESS FOR ANY PARTICULAR OR INTENDED PURPOSE, OR ANY OTHER WARRANTY OTHERWISE ARISING OUT OF ANY PROPOSAL, FINAL STANDARDS OR DRAFT STANDARDS, OR CONTRIBUTION.
Limitation of Liability
IN NO EVENT WILL CDISC OR ANY OF ITS CONSTITUENT PARTS (INCLUDING, BUT NOT LIMITED TO, THE CDISC BOARD OF DIRECTORS, THE CDISC PRESIDENT, CDISC STAFF, AND CDISC MEMBERS) BE LIABLE TO ANY OTHER PERSON OR ENTITY FOR ANY LOSS OF PROFITS, LOSS OF USE, DIRECT, INDIRECT, INCIDENTAL, CONSEQUENTIAL, OR SPECIAL DAMAGES, WHETHER UNDER CONTRACT, TORT, WARRANTY, OR OTHERWISE, ARISING IN ANY WAY OUT OF THIS POLICY OR ANY RELATED AGREEMENT, WHETHER OR NOT SUCH PARTY HAD ADVANCE NOTICE OF THE POSSIBILITY OF SUCH DAMAGES.
Note: The CDISC Intellectual Property Policy can be found at: cdisc_policy_003_intellectual_property_v201408.pdf.